Originally posted by Tukuler:
So what does your method reveal about say
* East Africa vs Tigara
or
* Pygmy vs EuroAmerican ?
By your method is Jebel Sahaba closer to
* East Africa
or
* West Africa?
What is the meaning of branches on the same limb?
What is being measured as the distance between populations?
quote:
Originally posted by the lioness,:
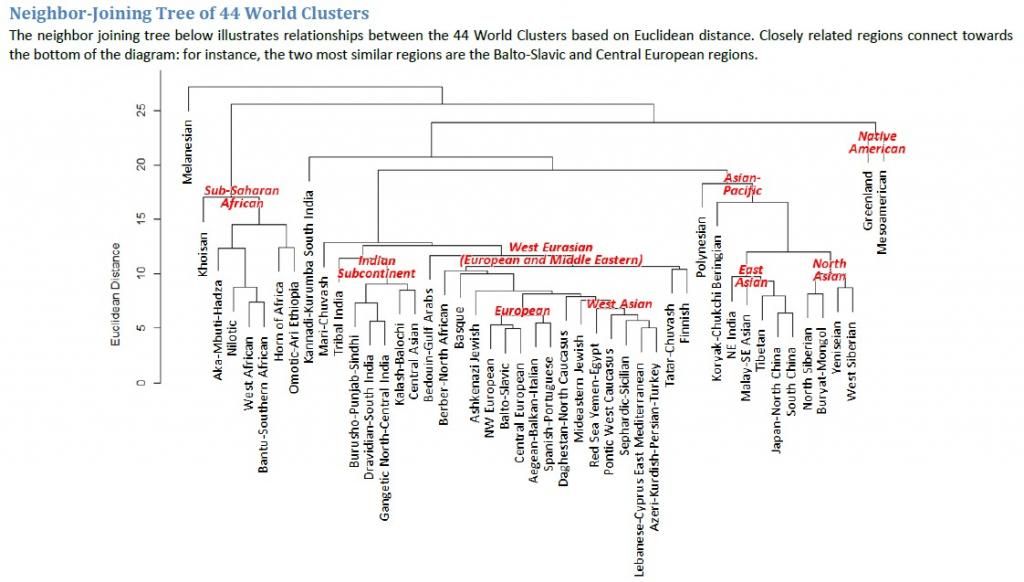
The way to determine the distance bewteen two populations on the above dendogram is to
start at the name of the first populaltion.
Then you follow the line path eminating from that name it like a mouse in a maze to the second popualation.
You can move in any direction along the path, up, down, backwards or forwards, whatever is the shortest distance from one population to the next.
Since the scale of this chart is horizontal you only record the distances of the horizontal movements.
When you put these horizontal segments together and measure them in total, that is the distance bewteen two populations






![[Confused]](confused.gif)