Originally posted by the lioness,:
quote:
Originally posted by Clyde Winters:
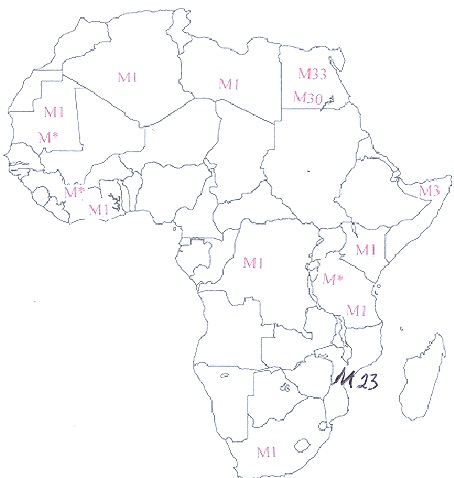
I believe most carriers of hg H migrated to Western Europe during the African invasion of Spain by Moors and Berbers and spread across Europe into the Middle East.
In summary, the presence of hg H in Europe is probably of recent origin.
The Tuareg and other Black Berber groups probably helped spread H in Europe after they invaded Europe along with other sahelians/Moors during the Islamic period.

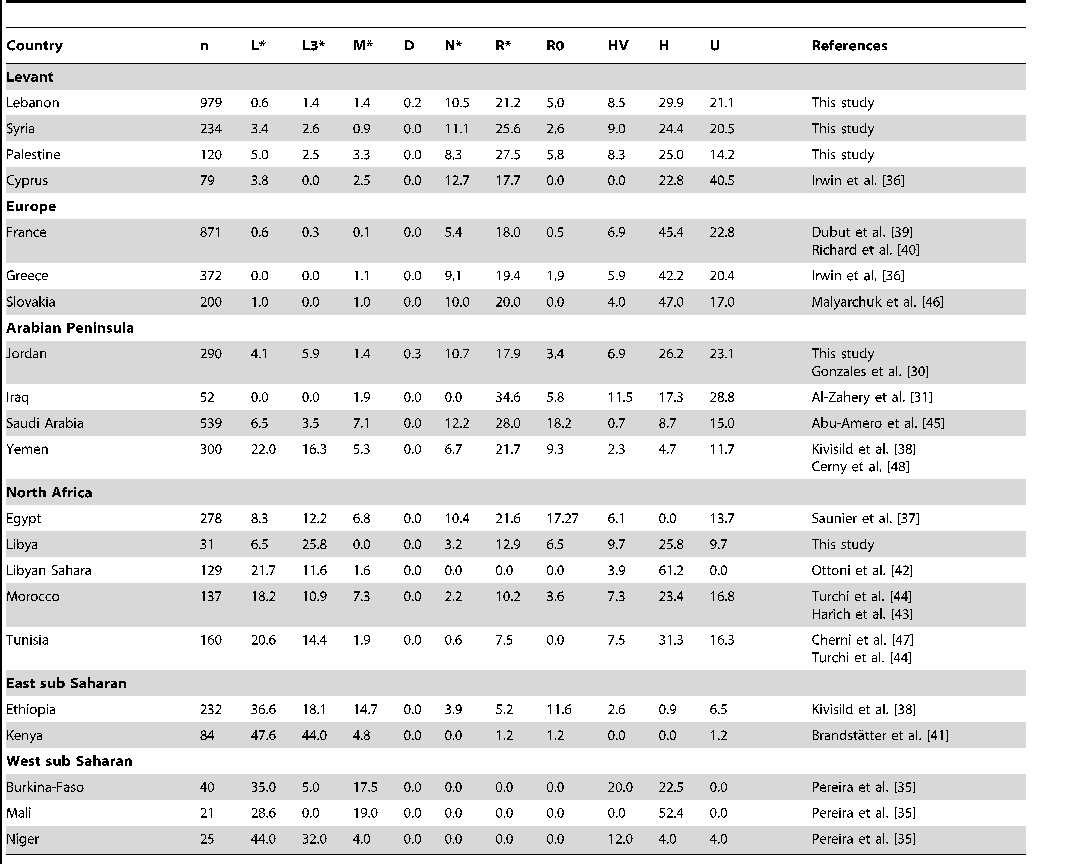
Clyde look at this modern day Germans are primarily African
Look
mtDNA
African H 44.8%
African U5 5.8%
Y DNA
African R1b about 40%
This proves that the Vandals have little or no relation to the modern day Germans














![[Big Grin]](biggrin.gif) at your rebuttal.
at your rebuttal. ![[Smile]](smile.gif)




![[Embarrassed]](redface.gif)

![[Wink]](wink.gif)



![[Roll Eyes]](rolleyes.gif)







