Originally posted by the lioness,:

Ancestors of R-V88 (ascending order)
M415
M343
M173
M207
quote:The Abstract points out that several related papers, i.e.
The cited paper (Winters, 2014) has avoided, unfortunately, an approval by the Editor-in-Chief, and slipped through to the Journal. The problem is that the paper contains several wrong and many highly questionable statements. As it happens with some authors, they selectively cite some references, often placing quotes out of context, ignore other references and data, which contradict their views. The above is a brief description of the cited article that is grossly misleading.
quote:.
This comment considers a paper by C. Winters recently published in Advances in Anthropology, as well as some of his related papers, concerning the “origin of haplogroup R1 in Africa”, “Neanderthals originated in Africa”, “Neanderthals spread from Morocco to East Africa”, “the Cro-Magnon people were probably Bushman/Khoisan”, and other inventions which do not belong to this Journal.
quote:The review ends with
Overall, the paper represents a collection of assorted quotations and declarations, many of them without any link to the title and/or the main subject of the paper. Most of declarations do not have links to any scientific reference, many of them reflect certain confusions, some references and data are taken out of context and described in a distorted manner.
Here is a typical example of the author’s style and reliability of quotations. In his preceding paper, entitled “Possible African origin of Y-chromosome R1-M173” (Winters, 2011) , the author writes: “The Khoisan also carry RM343 (R1b)… (Naidoo et al., 2010) the archaeological and linguistic data indicate the successful colonization of Asia by Sub-Saharan Africans from Nubia 5 - 4 kya”. If the reader looks up at the Naidoo et al. paper, it shows that the great majority of R1b-M343 haplotypes were found among South African Whites (81 out of 157), while Khoe-san contain three R1b-M343 haplotypes out of 183. Mr. Winters did not even mention such a discrepancy, which brings a different angle at the data.
Here is another example. In the same paper (Winters, 2011) he writes on “widespread distribution of R1*- M173 in Africa, that ranges between 7% - 95% and averages 39% (Coia et al., 2005) ”. He repeats the same “quotation” in yet another paper (Winters, 2010) ―“The frequency of Y-chromosome R1*-M173 in Africa range between 7% - 95% and averages 39.5% (Coia et al., 2005)”. However, in the referenced paper Coia et al. (2005) describe the frequency of R1*-M173 only in Cameroon, “with the highest frequency in North Cameroon (from 6.7% among the Tali to 95.2% among the Uldeme”. Mr. Winters did not mention that the figures were related not to “widespread distribution in Africa”, but specifically to North Cameroon, which is known since at least 2002(Cruciani et al., 2002, 2010) . In the same manner Mr. Winters “quote” data by Berniell-Lee et al. (2009) , who had reported that 5.2% of R1b1* were identified in Cameroon and neighboring Gabon, and “quote” it as follows “Haplogroup R1b1* is found in Africa…Berniell-Lee et al. (2005) found in their study that 5.2% carried Rb1*” (spelling by Winters, 2011 ). As one sees again, Cameroon and Gabon were not mentioned, only “found in Africa 5.2%…”.
Here is yet another example. According to Winters (2011) , “The bearers of R1b1* among the Pygmy populations ranged from 1% - 25% (Berniell-Lee et al., 2009) ”. In fact, Figure 1 in the Berniell-Lee et al. paper shows only two individuals having R1b1*, one from Baka tribe (out of 33 tested) in Gabon, and one from Bakola tribe (out of 22 tested) in Cameroon. So much for 1% - 25%. The list of misquotations can go on. That was a basis for the “African origin of R1-M173”.
Let us review an “age” of the R1b1* lineages in Cameroon and Gabon, based on haplotypes listed in (Berniell-Lee et al., 2009) . Figure 1 shows a haplotype tree, which is split into two distinct branches, on the left- and right-hand sides.
The right-hand side branch coalesces to the base (deduced ancestral) haplotype:
13 24 15 11 13 16 12 13 14 13 16 14 11 12 12 11 11 12 11
All 27 haplotypes of the branch contain 81 mutations from the base haplotype. It gives 81/27/0.0243 = 123 à 141 conditional generations (of 25 years each, see Klyosov, 2012 ); that is 3525 ± 530 years from a common ancestor of the branch. Here the arrow shows a correction for back mutations (Klyosov, 2009, 2012) , the margin of error calculated as described in (Klyosov, 2009) .
The left-hand side branch has the following base haplotype:
12 24 15 10 13 15 12 12 14 12 14 14 10 12 12 11 11 12 11
All 19 haplotypes of the branch contain 46 mutations. It gives 46/19/0.0243 = 100 à 111 conditional generations, that is 2775 ± 495 years from a common ancestor of the branch. One can see that the two populations could have migrated to Gabon about the same time (within the margin of error of the calculations), in the I-II millennium BC, and that time was not, of course, the time of “origin” of haplogroup R1. The latter arose around 26,000 years ago, in Central Asia, apparently in South Siberia (Klyosov, 2012) . Indeed, an excavated haplogroup R was recently identified in South Siberia (near the lake of Baikal), with an archaeological date of 24,000 years ago (Balter, 2013;Raghavan et al., 2013) .
In the same manner and style of misquotations and distorted “information”, Mr. Winters continues in his latest paper in Adv. Anthropol. (2014). He writes in the beginning of the paper―“Klyosov claims that the first Europeans were fair (pale) skin”, and “Klyosov argued that…ancient Europeans were fair (pale) skin and that most African haplogroups were the result of a back migration of pale skinned Europeans”. However, this is not the case in the paper (Klyosov, 2014) . The words “Europeans” and “skin” are never used in the paper next to each other, or in any other combination. There is nothing in the paper (Klyosov, 2014) regarding skin color of Europeans. There is nothing in the paper about “back migration of Europeans” to Africa, let alone “pale skinned Europeans”. Those are all inventions of Mr. Winters.
In other words, the paper (Klyosov, 2014) has nothing to do with the title of Mr. Winters article―“Were the first Europeans pale of dark skinned?” Apparently, Mr. Winters was seriously confused.
quote:
Indeed, the paper by Winters (2014) is grossly misleading
quote:--George B. J. Busby
New work [20–22] has addressed the Neolithic transition in Europe by focusing on the main western European Y chromosome haplogroup R1b1b2-M269 (hereafter referred to as R-M269). This lineage had hitherto received little recent attention in this context, although previous work suggested that the broader R-M173 clade (excluding the R1a-M17 sub-lineage) and Haplogroup 1 (derived at single nucleotide polymorphism, or SNP, 92r7) are likely to have spread into Europe during the Palaeolithic [17,18,23], and therefore unlikely to have been carried into Europe with the migrating farmers. Balaresque et al.
quote:--Chiara Batini
Sub-Saharan African Y chromosome diversity is represented by five main haplogroups (hgs): A, B, E, J, and R (Underhill et al. 2001; Cruciani et al. 2002; Tishkoff et al. 2007). Hgs J and R are geographically restricted to eastern and central Africa, respectively, whereas hg E shows a wider continental distribution (see also Berniell-Lee et al. 2009; Cruciani et al. 2010).
quote:--Fulvio Cruciani et al.
An independent high resolution MSY phylogeny has been recently obtained from 2,870 Y-SNPs discovered (or re- discovered) in the course of a large whole-genome re-sequencing study, but the observed variable sites all belong to the recent ‘‘out of Africa’’ CT clade [15]. Recently, in a re-sequencing study of the Y chromosome, the root of the tree moved to a new position and several changes at the basal nodes of the phylogeny were introduced [16]
[..]
Phylogenetic Mapping
Most of the mutations here analyzed belong to the African portion of the MSY phylogeny, which is comprised of haplogroups A1b, A1a, A2, A3 and B [16]. Through phylogenetic mapping it was possible to identify 15 new African haplogroups and to resolve one basal trifurcation (Figure 1). A new deep branch within the ‘‘out of Africa’’ haplogroup C was also identified (Figure S1).
Haplogroup A1b. The P114 mutation, which defines hap- logroup A1b according to Karafet et al. [14], had been detected in central-western Africa at very low frequencies (in total, three chromosomes from Cameroon) [16,19].
[...]
‘‘Out of Africa’’ haplogroups. All Y-clades that are not exclusively African belong to the macro-haplogroup CT, which is defined by mutations M168, M294 and P9.1 [14,31] and is subdivided into two major clades, DE and CF [1,14]. In a recent study [16], sequencing of two chromosomes belonging to haplogroups C and R, led to the identification of 25 new mutations, eleven of which were in the C-chromosome and seven in the R-chromosome. Here, the seven mutations which were found to be shared by chromosomes of haplogroups C and R [16], were also found to be present in one DE sample (sample 33 in Table S1), and positioned at the root of macro-haplogroup CT (Figure 1 and Figure S1). Six haplogroup C chromosomes (samples 34–39 in Table S1) were analyzed for the eleven haplogroup C- specific mutations [16] and for SNPs defining branches C1 to C6 in the tree by Karafet et al. [14] (Figure S1). Through this analysis we identified a chromosome from southern Europe as a new deep branch within haplogroup C (C-V20 or C7, Figure S1). Previously, only a few examples of C chromosomes (only defined by the marker RPS4Y711) had been found in southern Europe [32,33]. To improve our knowledge regarding the distribution of haplogroup C in Europe, we surveyed 1965 European subjects for the mutation RPS4Y711 and identified one additional haplogroup C chromosome from southern Europe, which has also been classified as C7 (data not shown). Further studies are needed to establish whether C7 chromosomes are the relics of an ancient European gene pool or the signal of a recent geographical spread from Asia. Two mutations, V248 and V87, which had never been previously described, were found to be specific to haplogroups C2 and C3, respectively (Figure S1). Three of the seven R-specific mutations (V45, V69 and V88) were previously mapped within haplogroup R [34], whereas the remaining four mutations have been here positioned at the root of haplogroups F (V186 and V205), K (V104) and P (V231) (Figure S1) through the analysis of 12 haplogroup F samples (samples 40–51, in Table S1).
[...]
Supporting Information
Figure S1 Structure of the macro-haplogroup CT. For details on mutations see legend to Figure 1. Dashed lines indicate putative branchings (no positive control available). The position of V248 (haplogroup C2) and V87 (haplogroup C3) compared to mutations that define internal branches was not determined. Note that mutations V45, V69 and V88 have been previously mapped (Cruciani et al. 2010; Eur J Hum Genet 18:800–807).
(TIF)
quote:--Fulvio Cruciani
does the present MSY tree compare with the backbone of the recently published “reference” MSY phylogeny?13 The phylogenetic relationships we observed among chromosomes belonging to haplogroups B, C, and R are reminiscent of those reported in the tree by Karafet et al.13
[...]
deepest branching separates A1b from a monophyletic clade whose members (A1a, A2, A3, B, C, and R) all share seven mutually reinforcing derived mutations (five transitions and two transversions, all at non-CpG sites).
[...]
The first branching in the MSY tree has been reported to be the one that separates the African-specific clade A (called clade I in 10) from clade BT (clade II-X in 10), whereas the second branching determines the subdivision of BT in clades B, mostly African, and CT, which comprises the majority of African and all non-African chromosomes.13,14 This branching pattern, along with the geographical distribu- tion of the major clades A, B, and CT, has been interpreted as supporting an African origin for anatomically modern humans,10 with Khoisan from south Africa and Ethiopians from east Africa sharing the deepest lineages of the phylogeny.15,16
[...]
To test the robustness of the backbone and the root of current Y chromosome phylogeny, we searched for SNPs that might be informative in this respect. To this aim, a resequencing analysis of a 205.9 kb MSY portion (183.5 kb in the X-degenerate and 22.4 kb in the X-transposed region) was performed for each of seven chromosomes that are representative of clade A (four chromosomes belonging to haplogroups A1a, A1b, A2, and A3), clade B, and clade CT (two chromosomes belonging to haplogroups C and R) (Table S1 available online).
The phylogenetic relationships we observed among chromosomes belonging to haplogroups B, C, and R are reminiscent of those reported in the tree by Karafet et al.13 These chromosomes belong to a clade (haplogroup BT) in which chromosomes C and R share a common ancestor (Figure 2).
quote:--Hong Shi et al. 2008:
The Y chromosome Alu polymorphism (YAP, also called M1) defines the deep-rooted haplogroup D/E of the global Y-chromosome phylogeny [1]. This D/E haplogroup is further branched into three sub-haplogroups DE*, D and E (Figure 1). The distribution of the D/E haplogroup is highly regional, and the three subgroups are geographically restricted to certain areas, therefore informative in tracing human prehistory (Table 1). The sub-haplogroup DE*, presumably the most ancient lineage of the D/E haplogroup was only found in Africans from Nigeria [2], supporting the "Out of Africa" hypothesis about modern human origin. The sub-haplogroup E (E-M40), defined by M40/SRY4064 and M96, was also suggested originated in Africa [3-6], and later dispersed to Middle East and Europe about 20,000 years ago [3,4]. Interestingly, the sub-haplogroup D defined by M174 (D-M174) is East Asian specific with abundant appearance in Tibetan and Japanese (30–40%), but rare in most of other East Asian populations and populations from regions bordering East Asia (Central Asia, North Asia and Middle East) (usually less than 5%) [5-7]. Under D-M174, Japanese belongs to a separate sub-lineage defined by several mutations (e.g. M55, M57 and M64 etc.), which is different from those in Tibetans implicating relatively deep divergence between them [1]. The fragmented distribution of D-M174 in East Asia seems not consistent with the pattern of other East Asian specific lineages, i.e. O3-M122, O1-M119 and O2-M95 under haplogroup O [8,9].
quote:Haplogroup DE* in Guinea-Bissau:
Further refinement awaits the finding of new markers especially within paragroup E3a*-M2. The microsatellite profile of the DE* individual is one mutational step away from the allelic state described for Nigerians (DYS390*21, DYS388 not tested; [37], therefore suggesting a common ancestry but not elucidating the phylogenetics.
quote:Haplogroup DE* in Nigerians:
There has been considerable debate on the geographic origin of the human Y chromosome Alu polymor- phism (YAP). Here we report a new, very rare deep-rooting haplogroup within the YAP clade, together with data on other deep-rooting YAP clades. The new haplogroup, found so far in only five Nigerians, is the least-derived YAP haplogroup according to currently known binary markers. However, because the interior branching order of the Y chromosome genealogical tree remains unknown, it is impossible to impute the origin of the YAP clade with certainty. We discuss the problems presented by rare deep-rooting lineages for Y chromosome phylogeography.
quote:LOL. This paper did not demolish anything. He has not falsified my research. In fact he acknowledges that my statistics are correct. Although he criticized my paper, he failed to mention that V88 is older than M269. Moreover, the variety of African populations that carry R1 supported my contention that this haplogroup was widespread in Africa. This was supported by the varied and diversified African populations.
Originally posted by Quetzalcoatl:
[URL] http://file.scirp.org/Html/2-1590439_51973.htm[/URL]
Advances in Anthropology
Vol.04 No.04(2014), Article ID:51973,4 pages
10.4236/aa.2014.44024
"A Comment on the Paper: Were the First Europeans Pale or Dark Skinned? (by C. Winters, Advances in Anthropology, 2014, 4, 124-132)"
Finally, a peer review of a paper by Clyde Winters, though the journal is Open Access it has peer review unlike most of the journals Winter’s publishes in. The reviewer would have recommended not publishing.
quote:The Abstract points out that several related papers, i.e.
The cited paper (Winters, 2014) has avoided, unfortunately, an approval by the Editor-in-Chief, and slipped through to the Journal. The problem is that the paper contains several wrong and many highly questionable statements. As it happens with some authors, they selectively cite some references, often placing quotes out of context, ignore other references and data, which contradict their views. The above is a brief description of the cited article that is grossly misleading.
Winters, C. (2010). The Kushite spread of Haplogroup R1*-M173 from Africa to Eurasia. Current Research Journal of Biological Sciences, 2, 294-299. and
Winters, C. (2011). Possible African origin of Y-chromosome R1-M173. International Journal of Science and Nature, 2, 743-745.
are also problematic.
quote:.
This comment considers a paper by C. Winters recently published in Advances in Anthropology, as well as some of his related papers, concerning the “origin of haplogroup R1 in Africa”, “Neanderthals originated in Africa”, “Neanderthals spread from Morocco to East Africa”, “the Cro-Magnon people were probably Bushman/Khoisan”, and other inventions which do not belong to this Journal.
The most relevant part concerns Winters’s claims about R1-M173
quote:The review ends with
Overall, the paper represents a collection of assorted quotations and declarations, many of them without any link to the title and/or the main subject of the paper. Most of declarations do not have links to any scientific reference, many of them reflect certain confusions, some references and data are taken out of context and described in a distorted manner.
Here is a typical example of the author’s style and reliability of quotations. In his preceding paper, entitled “Possible African origin of Y-chromosome R1-M173” (Winters, 2011) , the author writes: “The Khoisan also carry RM343 (R1b)… (Naidoo et al., 2010) the archaeological and linguistic data indicate the successful colonization of Asia by Sub-Saharan Africans from Nubia 5 - 4 kya”. If the reader looks up at the Naidoo et al. paper, it shows that the great majority of R1b-M343 haplotypes were found among South African Whites (81 out of 157), while Khoe-san contain three R1b-M343 haplotypes out of 183. Mr. Winters did not even mention such a discrepancy, which brings a different angle at the data.
Here is another example. In the same paper (Winters, 2011) he writes on “widespread distribution of R1*- M173 in Africa, that ranges between 7% - 95% and averages 39% (Coia et al., 2005) ”. He repeats the same “quotation” in yet another paper (Winters, 2010) ―“The frequency of Y-chromosome R1*-M173 in Africa range between 7% - 95% and averages 39.5% (Coia et al., 2005)”. However, in the referenced paper Coia et al. (2005) describe the frequency of R1*-M173 only in Cameroon, “with the highest frequency in North Cameroon (from 6.7% among the Tali to 95.2% among the Uldeme”. Mr. Winters did not mention that the figures were related not to “widespread distribution in Africa”, but specifically to North Cameroon, which is known since at least 2002(Cruciani et al., 2002, 2010) . In the same manner Mr. Winters “quote” data by Berniell-Lee et al. (2009) , who had reported that 5.2% of R1b1* were identified in Cameroon and neighboring Gabon, and “quote” it as follows “Haplogroup R1b1* is found in Africa…Berniell-Lee et al. (2005) found in their study that 5.2% carried Rb1*” (spelling by Winters, 2011 ). As one sees again, Cameroon and Gabon were not mentioned, only “found in Africa 5.2%…”.
Here is yet another example. According to Winters (2011) , “The bearers of R1b1* among the Pygmy populations ranged from 1% - 25% (Berniell-Lee et al., 2009) ”. In fact, Figure 1 in the Berniell-Lee et al. paper shows only two individuals having R1b1*, one from Baka tribe (out of 33 tested) in Gabon, and one from Bakola tribe (out of 22 tested) in Cameroon. So much for 1% - 25%. The list of misquotations can go on. That was a basis for the “African origin of R1-M173”.
Let us review an “age” of the R1b1* lineages in Cameroon and Gabon, based on haplotypes listed in (Berniell-Lee et al., 2009) . Figure 1 shows a haplotype tree, which is split into two distinct branches, on the left- and right-hand sides.
The right-hand side branch coalesces to the base (deduced ancestral) haplotype:
13 24 15 11 13 16 12 13 14 13 16 14 11 12 12 11 11 12 11
All 27 haplotypes of the branch contain 81 mutations from the base haplotype. It gives 81/27/0.0243 = 123 à 141 conditional generations (of 25 years each, see Klyosov, 2012 ); that is 3525 ± 530 years from a common ancestor of the branch. Here the arrow shows a correction for back mutations (Klyosov, 2009, 2012) , the margin of error calculated as described in (Klyosov, 2009) .
The left-hand side branch has the following base haplotype:
12 24 15 10 13 15 12 12 14 12 14 14 10 12 12 11 11 12 11
All 19 haplotypes of the branch contain 46 mutations. It gives 46/19/0.0243 = 100 à 111 conditional generations, that is 2775 ± 495 years from a common ancestor of the branch. One can see that the two populations could have migrated to Gabon about the same time (within the margin of error of the calculations), in the I-II millennium BC, and that time was not, of course, the time of “origin” of haplogroup R1. The latter arose around 26,000 years ago, in Central Asia, apparently in South Siberia (Klyosov, 2012) . Indeed, an excavated haplogroup R was recently identified in South Siberia (near the lake of Baikal), with an archaeological date of 24,000 years ago (Balter, 2013;Raghavan et al., 2013) .
In the same manner and style of misquotations and distorted “information”, Mr. Winters continues in his latest paper in Adv. Anthropol. (2014). He writes in the beginning of the paper―“Klyosov claims that the first Europeans were fair (pale) skin”, and “Klyosov argued that…ancient Europeans were fair (pale) skin and that most African haplogroups were the result of a back migration of pale skinned Europeans”. However, this is not the case in the paper (Klyosov, 2014) . The words “Europeans” and “skin” are never used in the paper next to each other, or in any other combination. There is nothing in the paper (Klyosov, 2014) regarding skin color of Europeans. There is nothing in the paper about “back migration of Europeans” to Africa, let alone “pale skinned Europeans”. Those are all inventions of Mr. Winters.
In other words, the paper (Klyosov, 2014) has nothing to do with the title of Mr. Winters article―“Were the first Europeans pale of dark skinned?” Apparently, Mr. Winters was seriously confused.quote:
Indeed, the paper by Winters (2014) is grossly misleading
quote:Klyosov wrote:
Abstract
This is an overview of the Out of Africa (OoA) settlement of Europe during the Aurignacian period.
Klyosov claims that the first Europeans were fair (pale) skin, and Neanderthal who never lived in Africa. Archaeological evidence indicated that Neanderthals originated in Africa and between 139 kya and 125 kya the Neanderthals migrated back into Africa and spread from Morocco to East Africa. The archaeological, anthropological and genetic evidence indicated that the first Europeans
were dark skin Sub-Saharan Africans who carried mtDNA haplogroup N and Y-chromosome C6 into
Europe.
Keywords
Haplogroup, Neanderthal, Phenotype, Skeletal, Mousterian, mtDNA, SLC24A5
quote:This was false as I noted in the introduction to my paper.
The words “Europeans” and “skin” are never used in the paper next to each other, or in any other combination. There is nothing in the paper (Klyosov, 2014) regarding skin color of Europeans.
quote:Although this is the opinion of Klyosov (2014) the archaeological and genetic evidence does not support his conclusion. The skeletal and archaeological evidence made it clear that the first Europeans were probably dark skin, not pale skin.”
“Ancestors of the most present-day non-Africans did not come from Africa in the last 30,000 - 600,000 years at least. In other words, those who migrated from Africa, or were forcefully taken out as slaves, are not ancestors of the contemporary Europeans, Asians, Native Americans, Australians, Polynesians. This follows from the whole multitude of data in anthropology, genetics, archaeology, DNA genealogy. Study of the DNA of excavated bones of Neanderthals has shown in them MCR1 melanocortin receptor, in the same variant as that in modern humans, which makes pale skin and red hair, observed in modern humans (Lalueza-Fox et al., 2007), though, according to the study authors, humans did not inherit MCR1 from Neanderthals. There was not any data that Neanderthals were Black Africans. Indeed, no Neanderthals were found in Africa.”
quote:Page 38
Strong waves of immigration have not been extensive until now, and the largest area of the island has been managed by the indigenous population through most of its human history – a recent molecular study (CERNY et al. 2009) showed that the Socotrans are not a genetic mixture of immigrants and local people, as previously believed.
quote:--Viktor Černý, Am J Phys Anthropol, 2009
The Soqotra archipelago is one of the most isolated landmasses in the world, situated at the mouth of the Gulf of Aden between the Horn of Africa and southern Arabia. The main island of Soqotra lies not far from the proposed southern migration route of anatomically modern humans out of Africa ∼60,000 years ago (kya), suggesting the island may harbor traces of that first dispersal. Nothing is known about the timing and origin of the first Soqotri settlers. The oldest historical visitors to the island in the 15th century reported only the presence of an ancient population. We collected samples throughout the island and analyzed mitochondrial DNA and Y-chromosomal variation. We found little African influence among the indigenous people of the island. Although the island population likely experienced founder effects, links to the Arabian Peninsula or southwestern Asia can still be found. In comparison with datasets from neighboring regions, the Soqotri population shows evidence of long-term isolation and autochthonous evolution of several mitochondrial haplogroups. Specifically, we identified two high-frequency founder lineages that have not been detected in any other populations and classified them as a new R0a1a1 subclade. Recent expansion of the novel lineages is consistent with a Holocene settlement of the island ∼6 kya.
quote:As usual. misdirection to avoid a difficulty. The reviewer was focused on your misquoting sources on R-M173 and dealt with several of your papers i.e.
Originally posted by Clyde Winters:
My paper was published in the same journal that published Klyosov (2014) i.e., Winters, C. (2014). Were the First Europeans Pale or Dark Skinned? Advances in Anthropology, 4,
124-132. http://dx.doi.org/10.4236/aa.2014.43016 . As a result, the article was peer reviewed. This paper was not about R1, as noted from the abstract.
quote:Klyosov wrote:
Abstract
This is an overview of the Out of Africa (OoA) settlement of Europe during the Aurignacian period.
Klyosov claims that the first Europeans were fair (pale) skin, and Neanderthal who never lived in Africa. Archaeological evidence indicated that Neanderthals originated in Africa and between 139 kya and 125 kya the Neanderthals migrated back into Africa and spread from Morocco to East Africa. The archaeological, anthropological and genetic evidence indicated that the first Europeans
were dark skin Sub-Saharan Africans who carried mtDNA haplogroup N and Y-chromosome C6 into
Europe.
Keywords
Haplogroup, Neanderthal, Phenotype, Skeletal, Mousterian, mtDNA, SLC24A5
quote:This was false as I noted in the introduction to my paper.
The words “Europeans” and “skin” are never used in the paper next to each other, or in any other combination. There is nothing in the paper (Klyosov, 2014) regarding skin color of Europeans.
“Introduction
Were the first Europeans dark or pale skinned? Klyosov (2014), argued that the Neanderthals and ancient Europeans were fair (pale) skin and that most African haplogroups were the result of a back migration of pale skinned Europeans. Klyosov (2014) claimed that archaeology and paleoanthropology data of African skeletal material did not tell us much about the origin of African and non-African populations. Finally, Klyosov (2014) argued that that there was no archaeological proof of the appearance of anatomically modern humans (AMH) in Africa dating before 100 kya. He wrote:quote:Although this is the opinion of Klyosov (2014) the archaeological and genetic evidence does not support his conclusion. The skeletal and archaeological evidence made it clear that the first Europeans were probably dark skin, not pale skin.”
“Ancestors of the most present-day non-Africans did not come from Africa in the last 30,000 - 600,000 years at least. In other words, those who migrated from Africa, or were forcefully taken out as slaves, are not ancestors of the contemporary Europeans, Asians, Native Americans, Australians, Polynesians. This follows from the whole multitude of data in anthropology, genetics, archaeology, DNA genealogy. Study of the DNA of excavated bones of Neanderthals has shown in them MCR1 melanocortin receptor, in the same variant as that in modern humans, which makes pale skin and red hair, observed in modern humans (Lalueza-Fox et al., 2007), though, according to the study authors, humans did not inherit MCR1 from Neanderthals. There was not any data that Neanderthals were Black Africans. Indeed, no Neanderthals were found in Africa.”
In the paper I illustrate that the first Europeans and Neanderthals were Black. He could not refute the evidence so he talked about R1.
.
quote:Socotrans
Originally posted by Troll Patrol # Ish Gebor:
quote:Page 38
Strong waves of immigration have not been extensive until now, and the largest area of the island has been managed by the indigenous population through most of its human history – a recent molecular study (CERNY et al. 2009) showed that the Socotrans are not a genetic mixture of immigrants and local people, as previously believed.
http://www.socotraproject.org/userfiles/files/Van%20Damme%20%20Banfield%202011%20Socotra%20Conservation.pdf
quote:--Viktor Černý, Am J Phys Anthropol, 2009
The Soqotra archipelago is one of the most isolated landmasses in the world, situated at the mouth of the Gulf of Aden between the Horn of Africa and southern Arabia. The main island of Soqotra lies not far from the proposed southern migration route of anatomically modern humans out of Africa ∼60,000 years ago (kya), suggesting the island may harbor traces of that first dispersal. Nothing is known about the timing and origin of the first Soqotri settlers. The oldest historical visitors to the island in the 15th century reported only the presence of an ancient population. We collected samples throughout the island and analyzed mitochondrial DNA and Y-chromosomal variation. We found little African influence among the indigenous people of the island. Although the island population likely experienced founder effects, links to the Arabian Peninsula or southwestern Asia can still be found. In comparison with datasets from neighboring regions, the Soqotri population shows evidence of long-term isolation and autochthonous evolution of several mitochondrial haplogroups. Specifically, we identified two high-frequency founder lineages that have not been detected in any other populations and classified them as a new R0a1a1 subclade. Recent expansion of the novel lineages is consistent with a Holocene settlement of the island ∼6 kya.
Out of Arabia—The settlement of Island Soqotra as revealed by mitochondrial and Y chromosome genetic diversity
http://onlinelibrary.wiley.com/doi/10.1002/ajpa.20960/abstract

quote:LOL. How could I have miss quoted any articles when I cited the populations that carried R1-M173. By citing the populations carrying the haplogroup I did not mislead anyone.
Originally posted by Quetzalcoatl:
quote:As usual. misdirection to avoid a difficulty. The reviewer was focused on your misquoting sources on R-M173 and dealt with several of your papers i.e.
Originally posted by Clyde Winters:
My paper was published in the same journal that published Klyosov (2014) i.e., Winters, C. (2014). Were the First Europeans Pale or Dark Skinned? Advances in Anthropology, 4,
124-132. http://dx.doi.org/10.4236/aa.2014.43016 . As a result, the article was peer reviewed. This paper was not about R1, as noted from the abstract.
quote:Klyosov wrote:
Abstract
This is an overview of the Out of Africa (OoA) settlement of Europe during the Aurignacian period.
Klyosov claims that the first Europeans were fair (pale) skin, and Neanderthal who never lived in Africa. Archaeological evidence indicated that Neanderthals originated in Africa and between 139 kya and 125 kya the Neanderthals migrated back into Africa and spread from Morocco to East Africa. The archaeological, anthropological and genetic evidence indicated that the first Europeans
were dark skin Sub-Saharan Africans who carried mtDNA haplogroup N and Y-chromosome C6 into
Europe.
Keywords
Haplogroup, Neanderthal, Phenotype, Skeletal, Mousterian, mtDNA, SLC24A5
quote:This was false as I noted in the introduction to my paper.
The words “Europeans” and “skin” are never used in the paper next to each other, or in any other combination. There is nothing in the paper (Klyosov, 2014) regarding skin color of Europeans.
“Introduction
Were the first Europeans dark or pale skinned? Klyosov (2014), argued that the Neanderthals and ancient Europeans were fair (pale) skin and that most African haplogroups were the result of a back migration of pale skinned Europeans. Klyosov (2014) claimed that archaeology and paleoanthropology data of African skeletal material did not tell us much about the origin of African and non-African populations. Finally, Klyosov (2014) argued that that there was no archaeological proof of the appearance of anatomically modern humans (AMH) in Africa dating before 100 kya. He wrote:quote:Although this is the opinion of Klyosov (2014) the archaeological and genetic evidence does not support his conclusion. The skeletal and archaeological evidence made it clear that the first Europeans were probably dark skin, not pale skin.”
“Ancestors of the most present-day non-Africans did not come from Africa in the last 30,000 - 600,000 years at least. In other words, those who migrated from Africa, or were forcefully taken out as slaves, are not ancestors of the contemporary Europeans, Asians, Native Americans, Australians, Polynesians. This follows from the whole multitude of data in anthropology, genetics, archaeology, DNA genealogy. Study of the DNA of excavated bones of Neanderthals has shown in them MCR1 melanocortin receptor, in the same variant as that in modern humans, which makes pale skin and red hair, observed in modern humans (Lalueza-Fox et al., 2007), though, according to the study authors, humans did not inherit MCR1 from Neanderthals. There was not any data that Neanderthals were Black Africans. Indeed, no Neanderthals were found in Africa.”
In the paper I illustrate that the first Europeans and Neanderthals were Black. He could not refute the evidence so he talked about R1.
.
"A Comment on the Paper: Were the First Europeans Pale or Dark Skinned? (by C. Winters, Advances in Anthropology, 2014, 4, 124-132)"
Winters, C. (2010). The Kushite spread of Haplogroup R1*-M173 from Africa to Eurasia. Current Research Journal of Biological Sciences, 2, 294-299. and
Winters, C. (2011). Possible African origin of Y-chromosome R1-M173. International Journal of Science and Nature, 2, 743-745.
quote:As usual. misdirection to avoid a difficulty. The reviewer was focused on your misquoting sources on R-M173 and dealt with several of your papers i.e.
Originally posted by Clyde Winters:
In the paper I illustrate that the first Europeans and Neanderthals were Black. He could not refute the evidence so he talked about R1.
.
quote:But , of course, you misquoted the papers you cited. You argue that R-M-173 is widespread in Africa, that's the main
LOL. How could I have miss quoted any articles when I cited the populations that carried R1-M173. By citing the populations carrying the haplogroup I did not mislead anyone.
.

quote:First, the reviewer of your article was anonymous ,chosen by the editor of the Journal, not Klysov. And, contrary to what you said that the reviewer could not deal with your Neanderthal stuff-- he did as well as some more on your R-M173 as follows:
Originally posted by Clyde Winters:
quote:]LOL. How could I have miss quoted any articles when I cited the populations that carried R1-M173. By citing the populations carrying the haplogroup I did not mislead anyone.
Originally posted by Clyde Winters:
[qb] My paper was published in the same journal that published Klyosov (2014) i.e., Winters, C. (2014). Were the First Europeans Pale or Dark Skinned? Advances in Anthropology, 4,
124-132. http://dx.doi.org/10.4236/aa.2014.43016 . As a result, the article was peer reviewed. This paper was not about R1, as noted from the abstract.
quote:Klyosov wrote:
Abstract
This is an overview of the Out of Africa (OoA) settlement of Europe during the Aurignacian period.
Klyosov claims that the first Europeans were fair (pale) skin, and Neanderthal who never lived in Africa. Archaeological evidence indicated that Neanderthals originated in Africa and between 139 kya and 125 kya the Neanderthals migrated back into Africa and spread from Morocco to East Africa. The archaeological, anthropological and genetic evidence indicated that the first Europeans
were dark skin Sub-Saharan Africans who carried mtDNA haplogroup N and Y-chromosome C6 into
Europe.
Keywords
Haplogroup, Neanderthal, Phenotype, Skeletal, Mousterian, mtDNA, SLC24A5
quote:This was false as I noted in the introduction to my paper.
The words “Europeans” and “skin” are never used in the paper next to each other, or in any other combination. There is nothing in the paper (Klyosov, 2014) regarding skin color of Europeans.
“Introduction
Were the first Europeans dark or pale skinned? Klyosov (2014), argued that the Neanderthals and ancient Europeans were fair (pale) skin and that most African haplogroups were the result of a back migration of pale skinned Europeans. Klyosov (2014) claimed that archaeology and paleoanthropology data of African skeletal material did not tell us much about the origin of African and non-African populations. Finally, Klyosov (2014) argued that that there was no archaeological proof of the appearance of anatomically modern humans (AMH) in Africa dating before 100 kya. He wrote:quote:Although this is the opinion of Klyosov (2014) the archaeological and genetic evidence does not support his conclusion. The skeletal and archaeological evidence made it clear that the first Europeans were probably dark skin, not pale skin.”
“Ancestors of the most present-day non-Africans did not come from Africa in the last 30,000 - 600,000 years at least. In other words, those who migrated from Africa, or were forcefully taken out as slaves, are not ancestors of the contemporary Europeans, Asians, Native Americans, Australians, Polynesians. This follows from the whole multitude of data in anthropology, genetics, archaeology, DNA genealogy. Study of the DNA of excavated bones of Neanderthals has shown in them MCR1 melanocortin receptor, in the same variant as that in modern humans, which makes pale skin and red hair, observed in modern humans (Lalueza-Fox et al., 2007), though, according to the study authors, humans did not inherit MCR1 from Neanderthals. There was not any data that Neanderthals were Black Africans. Indeed, no Neanderthals were found in Africa.”
In the paper I illustrate that the first Europeans and Neanderthals were Black. He could not refute the evidence so he talked about R1.
.
.
quote:
In the same manner and style of misquotations and distorted “information”, Mr. Winters continues in his latest paper in Adv. Anthropol. (2014). He writes in the beginning of the paper―“Klyosov claims that the first Europeans were fair (pale) skin”, and “Klyosov argued that…ancient Europeans were fair (pale) skin and that most African haplogroups were the result of a back migration of pale skinned Europeans”. However, this is not the case in the paper (Klyosov, 2014) . The words “Europeans” and “skin” are never used in the paper next to each other, or in any other combination. There is nothing in the paper (Klyosov, 2014) regarding skin color of Europeans. There is nothing in the paper about “back migration of Europeans” to Africa, let alone “pale skinned Europeans”. Those are all inventions of Mr. Winters.
In other words, the paper (Klyosov, 2014) has nothing to do with the title of Mr. Winters article―“Were the first Europeans pale of dark skinned?” Apparently, Mr. Winters was seriously confused. However, Mr. Winters has made several other statements, some of them worth consideration in this Comment; seven of them are as follows:
1) “Archaeological evidence indicated that Neanderthals originated in Africa” (no reference given).
Figure 1. The tree is composed of 46 of 19-marker haplotypes of haplogroup R1b1*, 44 of them from Gabon, and two (the Pygmy individuals) from Cameroon (both are on the right-hand side). The haplotype tree was composed using software PHYLIP, Phylogeny Inference Package program (Felsenstein, 2004) and MEGA, Molecular Evolutionary Genetics Analysis, Version 6.0 (Tamura et al., 2013) . The haplotypes employ the following markers: DYS 393, 390, 19, 391, 385a, 385b, 388, 439, 389-1, 393, 389-2, 437, 460, 438, 436, 462, 434, 461, 435.
2) “Between 139 and 125 kya the Neanderthals migrated back into Africa and spread from Morocco to East Africa (Ki-Zerbo, 1981: p. 572)”.
3) “The archaeological, anthropological and genetic evidence indicated that the first Europeans were dark skin Sub-Saharan Africans” (no reference given).
4) “Sub-Saharan Africans…carried Y-chromosome C6 into Europe” (no reference given).
5) “The archaeological research makes it clear Neanderthal probably mixed with Africans” (no reference given).
6) “The Cro-Magnon people were probably Bushman/Khoisan (Boule & Vallois, 1957) ”.
7) “The archaeological and craniometric evidence indicates that the pre-Indo-European people were probably highly pigmented” (no reference given).
Let me explain:
Item 1. I would be delighted to learn of that “archaeological evidence”, that “Neanderthals originated in Africa”. However, alas, there was no reference provided. On the contrary, there are sufficient references that essentially repeat this: “Neanderthals, Homo Neanderthalensis, lived between 350,000 and 24,500 ya, throughout Europe and the Middle East, but…no Neanderthals fossils have yet been found in Africa” (Fuerle, 2008) .
Here is an another quotation, from a paper on genome studies: “Our analysis of human-Neandertal data…confirms previous claims that Neandertals contributed genetically to contemporary Eurasian populations (Green et al., 2010; Sankararaman et al., 2012; Yang et al., 2012) . However, in contrast to previous studies we can conclusively reject long-term population structure in the ancestral African population as an alternative explanation for the excess sharing of derived mutations by Neandertals and Eurasians” (Lohse & Frantz, 2014) .
Since Mr. Winters often cites the book “Fossil Men” by Boule and Vallois (1957) , it would be appropriate to note that in the sizeable chapter “Neandertal Man” (pp. 193-258) the word “Africa” is mentioned only once, in the Conclusions section, in a negative context, that Neanderthals are “very different…from the Eskimoes, the Fuegians, the Bushmen, the Pygmies, African or Asiatic, the Veddas, the Polynesians, the Melanesians, and even from the Australians, with whom attempts at comparison have often been made” (p. 252).
Item 2. Comments in Item 1 show that “spread from Morocco to East Africa” for Neanderthals after “between 139 and 125 kya” is not supported by any archaeological or other data.
Item 3. The archaeological and anthropological” evidence cannot witness the “dark skin” color of “the first Europeans”, much less that they were “Sub-Saharan Africans”. No wonder that no reference was provided. Regarding genetic data, all that they have shown is that “both Loschbour and Stuttgart had dark hair…and Loschbour, like La Braña and Motala12, probably had blue or light colored eyes…whereas Stuttgart probably had brown eyes. Neither Loschbour nor La Braña carries the skin-lightening allele in SLC24A5…” (Lazaridis et al., 2014) . They were not “the first Europeans”, they lived 7000 - 8000 ya. Regarding the first Europeans, there is no genetic data available, and archaeological data on excavated bones do not (and cannot) say anything on their skin color.
Item 4. There is no data that haplogroup/subclade C6 was brought to Europe by “Sub-Saharan Africans”. No wonder that a reference was not provided.
Item 5. There is no archaeological research which “makes it clear” that “Neanderthal probably mixed with Africans”. Neither “clear”, nor “probably”. Genetics does not show such a connection(Vernot & Akey, 2014; Reyes-Centeno et al., 2014; Sankararaman et al., 2014; Lohse & Frantz, 2014) .
Item 6. The statement that “The Cro-Magnon people were probably Bushman/Khoisan” cannot be considered seriously without more direct data than a reference related to 1915 as “Peringuey has told us that in certain burials on the South African coast ‘associated with the Aurignacian or Solutrean type of industry” (Boule & Vallois, 1957: p. 319) .
Item 7. There is no “archaeological and craniometric evidence” that can possibly indicate that “the pre-In- do-European people were probably highly pigmented”. Again, no wonder that a reference was not provided.
Indeed, the paper by Winters (2014) is grossly misleading.
References
1. Balter, M. (2013) . Ancient DNA links Native Americans with Europe. Science, 342, 409-410. http://dx.doi.org/10.1126/science.342.6157.409
2. Berniell-Lee, G., Calafell, F., Bosch, E., Heyer, E., Sica, L., Mouguiama-Daouda, P., van der Veen, L., Hombert, J.-M., et al. (2009). Genetic and Demographic Implications of the Bantu Expansion: Insights from Human Paternal Lineages. Molecular Biology and Evolution, 26, 1581-1589. http://dx.doi.org/10.1093/molbev/msp069
3. Boule, M., & Vallois, H. V. (1957). Fossil Men. The Dryden Press, New York, 535 p.
4. Coia, V., Destro-Bisol, G., Verginelli, F., Battagia, C., Boschi, I., Cruciani, F., Spedini, G., Comas, D., & Calafell, F. (2005). Brief Communication: mtDNA Variation in North Cameroon: Lack of Asian Lineages and Implications for Back Migration from Asia to Sub-Saharan Africa. American Journal of Physical Anthropology, 128, 678-681.http://dx.doi.org/10.1002/ajpa.20138
5. Cruciani, F., Santolamazza, P., Shen, P., Makaulay, V., Moral, P., Olckers. A., Modiano, D., Holmes, S., et al. (2002). A back Migration from Asia to Sub-Saharan Africa Is Supported by High-Resolution Analysis of Human Y-Chromosome Haplotypes. The American Journal of Human Genetics, 70, 1197-1214. http://dx.doi.org/10.1086/340257
6. Cruciani, F., Trombetta, B., Sellitto, D., Massaia, A., Destro-Bisol, G., Watson, E., Colomb, B. E., Dugoujon, J. M., Moral, P., & Scozzari, R. (2010). Human Y Chromosome Haplogroup R-V88: A Paternal Genetic Record of Early Mid Holocene Trans-Saharan Connections and the Spread of Chadic Languages. European Journal of Human Genetics, 18, 800-807. http://dx.doi.org/10.1038/ejhg.2009.231
7. Felsenstein, J. (2004). PHYLIP (Phylogeny Inference Package). Seattle, WA: Department of Genome Sciences, University of Washington.
8. Fuerle, R. D. (2008). Erectus Walks among Us. New York: Spooner Press, 340 p.
9. Green, R. E., Krause, J., Briggs, A. W., Maricic, T., Stenzel, U., et al. (2010). A Draft Sequence of the Neanderthal Genome. Science, 328, 710-722.
10. Klyosov, A . A. (2009). DNA Genealogy, Mutation Rates, and Some Historical Evidences Written in Y-Chromosome. I. Basic Principles and the Method. Journal of Genetic Genealogy, 5, 186-216.
11. Klyosov, A. A. (2012). Ancient History of the Arbins, Bearers of Haplogroup R1b, from Central Asia to Europe, 16,000 to 1500 Years before Present. Advances in Anthropology, 2, 87-105. http://dx.doi.org/10.4236/aa.2012.22010
12. Klyosov, A. A. (2014). Reconsideration of the “Out of Africa” Concept as Not Having Enough Proof. Advances in Anthropology, 4, 18-37.http://dx.doi.org/10.4236/aa.2014.41004
13. Lazaridis, I., Patterson, N., Mittnik, A., Renaud, G., Mallick, S., Kirsanow, K., Sudmant, P. H., Schraiber, J. G., et al. (2014) Ancient Human Genomes Suggest Three Ancestral Populations for Present-Day Europeans. Nature, 513, 409-413.http://dx.doi.org/10.1038/nature13673
14. Lohse, K., & Frantz, L. A. F. (2014). Neandertal Admixture in Eurasia Confirmed by Maximum-Likelihood Analysis of Three Genomes. Genetics, 196, 1241-1251.http://dx.doi.org/10.1534/genetics.114.162396
15. Naidoo, T., Schlebusch, C. M., Makkan, H., Patel, P., Mahabeer, R., Erasmus, J. C., & Soodyall, H. (2010). Development of a Single Base Extension Method to Resolve Y Chromosome Haplogroups in Sub-Saharan African Populations. Investigative Genetics, 1, 1-11. http://dx.doi.org/10.1186/2041-2223-1-6
16. Raghavan, M., Skoglund, P., Graf, K. E., Metspalu, M., Albrechtsen, A., Moltke, I., et al. (2013). Upper Palaeolithic Siberian Genome Reveals Dual Ancestry of Native Americans. Nature, 505, 87-91. http://dx.doi.org/10.1038/nature12736
17. Reyes-Centeno, H., Ghirotto, S., Detroit, F., Grimaud-Herve, D., Barbujani, G., & Harvati, K. (2014). Genomic and Cranial Phenotype Data Support Multiple Modern Human Dispersals from Africa and a Southern Route into Asia. Proceedings of the National Academy of Sciences of the United States of America, 111, 7248-7253.http://dx.doi.org/10.1073/pnas.1323666111
18. Sankararaman, S., Mallick, S., Dannemann, M., Prüfer, K., Kelso, J., Pääbo, S., Patterson, N., & Reich, D. (2014). The Genomic Landscape of Neanderthal Ancestry in Present-Day Humans. Nature, 507, 354-357. http://dx.doi.org/10.1038/nature12961
19. Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution, 30, 2725-2729. http://dx.doi.org/10.1093/molbev/mst197
20. Vernot, B., & Akey, J. M. (2014). Resurrecting Surviving Neandertal Lineages from Modern Human Genomes. Science, 343, 1017-1021.http://dx.doi.org/10.1126/science.1245938
21. Winters, C. (2010). The Kushite spread of Haplogroup R1*-M173 from Africa to Eurasia. Current Research Journal of Biological Sciences, 2, 294-299.
22. Winters, C. (2011). Possible African origin of Y-chromosome R1-M173. International Journal of Science and Nature, 2, 743-745.
23. Winters, C. (2014). Were the First Europeans Pale or Dark Skinned? Advances in Anthropology, 4, 124-132. http://dx.doi.org/10.4236/aa.2014.43016
24. Yang, M. A., Malaspinas, A.-S., Durand, E. Y., & Slatkin, M. (2012). Ancient Structure in Africa Unlikely to Explain Neanderthal and Non-African Genetic Similarity. Molecular Biology and Evolution, 29, 2987-2995. http://dx.doi.org/10.1093/molbev/mss117
quote:
Originally posted by the lioness,:
Ancestors of R-V88 (ascending order)
M415
M343
M173
M207
quote:
Originally posted by the lioness,:
Ancestors of R-V88 (ascending order)
M415
M343
M173
M207
quote:maybe you didn't read the post
Originally posted by xyyman:
Exactly my point. European L11 is, what, 5 mutational steps below R-V88
quote:the only subclade of R-V88 is
Originally posted by xyyman:
European L11 is, what, 5 mutational steps below R-V88
quote:This paper as I said earlier does not falsify anything. He was able to dispute the research that shows the first Europeans were not pale,he could not dispute the fact that neither Loschbour nor La Braña carries the skin-lightening allele in SLC24A5…” (Lazaridis et al., 2014), nor was he able to show that the Neanderthal were pale-skinned. Moreover, he did not cite any articles that dispute Boule & Vallois, 1957 research that the Khoisan were the first Europeans.
Originally posted by Quetzalcoatl:
quote:First, the reviewer of your article was anonymous ,chosen by the editor of the Journal, not Klysov. And, contrary to what you said that the reviewer could not deal with your Neanderthal stuff-- he did as well as some more on your R-M173 as follows:
Originally posted by Clyde Winters:
quote:]LOL. How could I have miss quoted any articles when I cited the populations that carried R1-M173. By citing the populations carrying the haplogroup I did not mislead anyone.
Originally posted by Clyde Winters:
[qb] My paper was published in the same journal that published Klyosov (2014) i.e., Winters, C. (2014). Were the First Europeans Pale or Dark Skinned? Advances in Anthropology, 4,
124-132. http://dx.doi.org/10.4236/aa.2014.43016 . As a result, the article was peer reviewed. This paper was not about R1, as noted from the abstract.
quote:Klyosov wrote:
Abstract
This is an overview of the Out of Africa (OoA) settlement of Europe during the Aurignacian period.
Klyosov claims that the first Europeans were fair (pale) skin, and Neanderthal who never lived in Africa. Archaeological evidence indicated that Neanderthals originated in Africa and between 139 kya and 125 kya the Neanderthals migrated back into Africa and spread from Morocco to East Africa. The archaeological, anthropological and genetic evidence indicated that the first Europeans
were dark skin Sub-Saharan Africans who carried mtDNA haplogroup N and Y-chromosome C6 into
Europe.
Keywords
Haplogroup, Neanderthal, Phenotype, Skeletal, Mousterian, mtDNA, SLC24A5
quote:This was false as I noted in the introduction to my paper.
The words “Europeans” and “skin” are never used in the paper next to each other, or in any other combination. There is nothing in the paper (Klyosov, 2014) regarding skin color of Europeans.
“Introduction
Were the first Europeans dark or pale skinned? Klyosov (2014), argued that the Neanderthals and ancient Europeans were fair (pale) skin and that most African haplogroups were the result of a back migration of pale skinned Europeans. Klyosov (2014) claimed that archaeology and paleoanthropology data of African skeletal material did not tell us much about the origin of African and non-African populations. Finally, Klyosov (2014) argued that that there was no archaeological proof of the appearance of anatomically modern humans (AMH) in Africa dating before 100 kya. He wrote:quote:Although this is the opinion of Klyosov (2014) the archaeological and genetic evidence does not support his conclusion. The skeletal and archaeological evidence made it clear that the first Europeans were probably dark skin, not pale skin.”
“Ancestors of the most present-day non-Africans did not come from Africa in the last 30,000 - 600,000 years at least. In other words, those who migrated from Africa, or were forcefully taken out as slaves, are not ancestors of the contemporary Europeans, Asians, Native Americans, Australians, Polynesians. This follows from the whole multitude of data in anthropology, genetics, archaeology, DNA genealogy. Study of the DNA of excavated bones of Neanderthals has shown in them MCR1 melanocortin receptor, in the same variant as that in modern humans, which makes pale skin and red hair, observed in modern humans (Lalueza-Fox et al., 2007), though, according to the study authors, humans did not inherit MCR1 from Neanderthals. There was not any data that Neanderthals were Black Africans. Indeed, no Neanderthals were found in Africa.”
In the paper I illustrate that the first Europeans and Neanderthals were Black. He could not refute the evidence so he talked about R1.
.
.
quote:
In the same manner and style of misquotations and distorted “information”, Mr. Winters continues in his latest paper in Adv. Anthropol. (2014). He writes in the beginning of the paper―“Klyosov claims that the first Europeans were fair (pale) skin”, and “Klyosov argued that…ancient Europeans were fair (pale) skin and that most African haplogroups were the result of a back migration of pale skinned Europeans”. However, this is not the case in the paper (Klyosov, 2014) . The words “Europeans” and “skin” are never used in the paper next to each other, or in any other combination. There is nothing in the paper (Klyosov, 2014) regarding skin color of Europeans. There is nothing in the paper about “back migration of Europeans” to Africa, let alone “pale skinned Europeans”. Those are all inventions of Mr. Winters.
In other words, the paper (Klyosov, 2014) has nothing to do with the title of Mr. Winters article―“Were the first Europeans pale of dark skinned?” Apparently, Mr. Winters was seriously confused. However, Mr. Winters has made several other statements, some of them worth consideration in this Comment; seven of them are as follows:
1) “Archaeological evidence indicated that Neanderthals originated in Africa” (no reference given).
Figure 1. The tree is composed of 46 of 19-marker haplotypes of haplogroup R1b1*, 44 of them from Gabon, and two (the Pygmy individuals) from Cameroon (both are on the right-hand side). The haplotype tree was composed using software PHYLIP, Phylogeny Inference Package program (Felsenstein, 2004) and MEGA, Molecular Evolutionary Genetics Analysis, Version 6.0 (Tamura et al., 2013) . The haplotypes employ the following markers: DYS 393, 390, 19, 391, 385a, 385b, 388, 439, 389-1, 393, 389-2, 437, 460, 438, 436, 462, 434, 461, 435.
2) “Between 139 and 125 kya the Neanderthals migrated back into Africa and spread from Morocco to East Africa (Ki-Zerbo, 1981: p. 572)”.
3) “The archaeological, anthropological and genetic evidence indicated that the first Europeans were dark skin Sub-Saharan Africans” (no reference given).
4) “Sub-Saharan Africans…carried Y-chromosome C6 into Europe” (no reference given).
5) “The archaeological research makes it clear Neanderthal probably mixed with Africans” (no reference given).
6) “The Cro-Magnon people were probably Bushman/Khoisan (Boule & Vallois, 1957) ”.
7) “The archaeological and craniometric evidence indicates that the pre-Indo-European people were probably highly pigmented” (no reference given).
Let me explain:
Item 1. I would be delighted to learn of that “archaeological evidence”, that “Neanderthals originated in Africa”. However, alas, there was no reference provided. On the contrary, there are sufficient references that essentially repeat this: “Neanderthals, Homo Neanderthalensis, lived between 350,000 and 24,500 ya, throughout Europe and the Middle East, but…no Neanderthals fossils have yet been found in Africa” (Fuerle, 2008) .
Here is an another quotation, from a paper on genome studies: “Our analysis of human-Neandertal data…confirms previous claims that Neandertals contributed genetically to contemporary Eurasian populations (Green et al., 2010; Sankararaman et al., 2012; Yang et al., 2012) . However, in contrast to previous studies we can conclusively reject long-term population structure in the ancestral African population as an alternative explanation for the excess sharing of derived mutations by Neandertals and Eurasians” (Lohse & Frantz, 2014) .
Since Mr. Winters often cites the book “Fossil Men” by Boule and Vallois (1957) , it would be appropriate to note that in the sizeable chapter “Neandertal Man” (pp. 193-258) the word “Africa” is mentioned only once, in the Conclusions section, in a negative context, that Neanderthals are “very different…from the Eskimoes, the Fuegians, the Bushmen, the Pygmies, African or Asiatic, the Veddas, the Polynesians, the Melanesians, and even from the Australians, with whom attempts at comparison have often been made” (p. 252).
Item 2. Comments in Item 1 show that “spread from Morocco to East Africa” for Neanderthals after “between 139 and 125 kya” is not supported by any archaeological or other data.
Item 3. The archaeological and anthropological” evidence cannot witness the “dark skin” color of “the first Europeans”, much less that they were “Sub-Saharan Africans”. No wonder that no reference was provided. Regarding genetic data, all that they have shown is that “both Loschbour and Stuttgart had dark hair…and Loschbour, like La Braña and Motala12, probably had blue or light colored eyes…whereas Stuttgart probably had brown eyes. Neither Loschbour nor La Braña carries the skin-lightening allele in SLC24A5…” (Lazaridis et al., 2014) . They were not “the first Europeans”, they lived 7000 - 8000 ya. Regarding the first Europeans, there is no genetic data available, and archaeological data on excavated bones do not (and cannot) say anything on their skin color.
Item 4. There is no data that haplogroup/subclade C6 was brought to Europe by “Sub-Saharan Africans”. No wonder that a reference was not provided.
Item 5. There is no archaeological research which “makes it clear” that “Neanderthal probably mixed with Africans”. Neither “clear”, nor “probably”. Genetics does not show such a connection(Vernot & Akey, 2014; Reyes-Centeno et al., 2014; Sankararaman et al., 2014; Lohse & Frantz, 2014) .
Item 6. The statement that “The Cro-Magnon people were probably Bushman/Khoisan” cannot be considered seriously without more direct data than a reference related to 1915 as “Peringuey has told us that in certain burials on the South African coast ‘associated with the Aurignacian or Solutrean type of industry” (Boule & Vallois, 1957: p. 319) .
Item 7. There is no “archaeological and craniometric evidence” that can possibly indicate that “the pre-In- do-European people were probably highly pigmented”. Again, no wonder that a reference was not provided.
Indeed, the paper by Winters (2014) is grossly misleading.
References
1. Balter, M. (2013) . Ancient DNA links Native Americans with Europe. Science, 342, 409-410. http://dx.doi.org/10.1126/science.342.6157.409
2. Berniell-Lee, G., Calafell, F., Bosch, E., Heyer, E., Sica, L., Mouguiama-Daouda, P., van der Veen, L., Hombert, J.-M., et al. (2009). Genetic and Demographic Implications of the Bantu Expansion: Insights from Human Paternal Lineages. Molecular Biology and Evolution, 26, 1581-1589. http://dx.doi.org/10.1093/molbev/msp069
3. Boule, M., & Vallois, H. V. (1957). Fossil Men. The Dryden Press, New York, 535 p.
4. Coia, V., Destro-Bisol, G., Verginelli, F., Battagia, C., Boschi, I., Cruciani, F., Spedini, G., Comas, D., & Calafell, F. (2005). Brief Communication: mtDNA Variation in North Cameroon: Lack of Asian Lineages and Implications for Back Migration from Asia to Sub-Saharan Africa. American Journal of Physical Anthropology, 128, 678-681.http://dx.doi.org/10.1002/ajpa.20138
5. Cruciani, F., Santolamazza, P., Shen, P., Makaulay, V., Moral, P., Olckers. A., Modiano, D., Holmes, S., et al. (2002). A back Migration from Asia to Sub-Saharan Africa Is Supported by High-Resolution Analysis of Human Y-Chromosome Haplotypes. The American Journal of Human Genetics, 70, 1197-1214. http://dx.doi.org/10.1086/340257
6. Cruciani, F., Trombetta, B., Sellitto, D., Massaia, A., Destro-Bisol, G., Watson, E., Colomb, B. E., Dugoujon, J. M., Moral, P., & Scozzari, R. (2010). Human Y Chromosome Haplogroup R-V88: A Paternal Genetic Record of Early Mid Holocene Trans-Saharan Connections and the Spread of Chadic Languages. European Journal of Human Genetics, 18, 800-807. http://dx.doi.org/10.1038/ejhg.2009.231
7. Felsenstein, J. (2004). PHYLIP (Phylogeny Inference Package). Seattle, WA: Department of Genome Sciences, University of Washington.
8. Fuerle, R. D. (2008). Erectus Walks among Us. New York: Spooner Press, 340 p.
9. Green, R. E., Krause, J., Briggs, A. W., Maricic, T., Stenzel, U., et al. (2010). A Draft Sequence of the Neanderthal Genome. Science, 328, 710-722.
10. Klyosov, A . A. (2009). DNA Genealogy, Mutation Rates, and Some Historical Evidences Written in Y-Chromosome. I. Basic Principles and the Method. Journal of Genetic Genealogy, 5, 186-216.
11. Klyosov, A. A. (2012). Ancient History of the Arbins, Bearers of Haplogroup R1b, from Central Asia to Europe, 16,000 to 1500 Years before Present. Advances in Anthropology, 2, 87-105. http://dx.doi.org/10.4236/aa.2012.22010
12. Klyosov, A. A. (2014). Reconsideration of the “Out of Africa” Concept as Not Having Enough Proof. Advances in Anthropology, 4, 18-37.http://dx.doi.org/10.4236/aa.2014.41004
13. Lazaridis, I., Patterson, N., Mittnik, A., Renaud, G., Mallick, S., Kirsanow, K., Sudmant, P. H., Schraiber, J. G., et al. (2014) Ancient Human Genomes Suggest Three Ancestral Populations for Present-Day Europeans. Nature, 513, 409-413.http://dx.doi.org/10.1038/nature13673
14. Lohse, K., & Frantz, L. A. F. (2014). Neandertal Admixture in Eurasia Confirmed by Maximum-Likelihood Analysis of Three Genomes. Genetics, 196, 1241-1251.http://dx.doi.org/10.1534/genetics.114.162396
15. Naidoo, T., Schlebusch, C. M., Makkan, H., Patel, P., Mahabeer, R., Erasmus, J. C., & Soodyall, H. (2010). Development of a Single Base Extension Method to Resolve Y Chromosome Haplogroups in Sub-Saharan African Populations. Investigative Genetics, 1, 1-11. http://dx.doi.org/10.1186/2041-2223-1-6
16. Raghavan, M., Skoglund, P., Graf, K. E., Metspalu, M., Albrechtsen, A., Moltke, I., et al. (2013). Upper Palaeolithic Siberian Genome Reveals Dual Ancestry of Native Americans. Nature, 505, 87-91. http://dx.doi.org/10.1038/nature12736
17. Reyes-Centeno, H., Ghirotto, S., Detroit, F., Grimaud-Herve, D., Barbujani, G., & Harvati, K. (2014). Genomic and Cranial Phenotype Data Support Multiple Modern Human Dispersals from Africa and a Southern Route into Asia. Proceedings of the National Academy of Sciences of the United States of America, 111, 7248-7253.http://dx.doi.org/10.1073/pnas.1323666111
18. Sankararaman, S., Mallick, S., Dannemann, M., Prüfer, K., Kelso, J., Pääbo, S., Patterson, N., & Reich, D. (2014). The Genomic Landscape of Neanderthal Ancestry in Present-Day Humans. Nature, 507, 354-357. http://dx.doi.org/10.1038/nature12961
19. Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution, 30, 2725-2729. http://dx.doi.org/10.1093/molbev/mst197
20. Vernot, B., & Akey, J. M. (2014). Resurrecting Surviving Neandertal Lineages from Modern Human Genomes. Science, 343, 1017-1021.http://dx.doi.org/10.1126/science.1245938
21. Winters, C. (2010). The Kushite spread of Haplogroup R1*-M173 from Africa to Eurasia. Current Research Journal of Biological Sciences, 2, 294-299.
22. Winters, C. (2011). Possible African origin of Y-chromosome R1-M173. International Journal of Science and Nature, 2, 743-745.
23. Winters, C. (2014). Were the First Europeans Pale or Dark Skinned? Advances in Anthropology, 4, 124-132. http://dx.doi.org/10.4236/aa.2014.43016
24. Yang, M. A., Malaspinas, A.-S., Durand, E. Y., & Slatkin, M. (2012). Ancient Structure in Africa Unlikely to Explain Neanderthal and Non-African Genetic Similarity. Molecular Biology and Evolution, 29, 2987-2995. http://dx.doi.org/10.1093/molbev/mss117
quote:
Originally posted by xyyman:
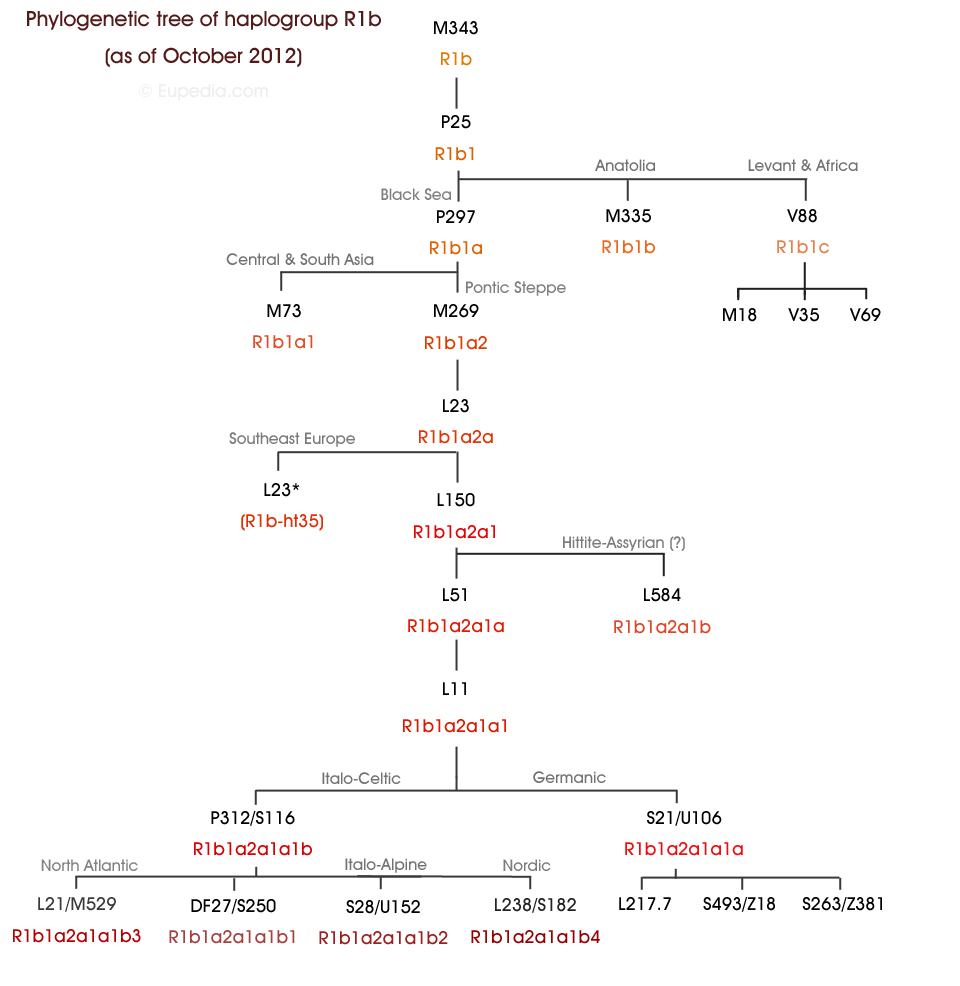
Oooookkkkk!! What paper states R1b has highest diversity OUTSIDE Africa cf to IN Africa?
Busby refused to peform or at least disclose a latitudinal analyis which would help resolve this. So all we have left is the Phylotree. So we are back to Wood et al.

quote:That is 100% WRONG
Originally posted by xyyman:
The Pontic Steppe is a sub-clade(downstream) of African R-V88.
quote:LOL at Low Level Thinker.
Originally posted by the lioness,:
Quiet jackass I'm not talking to you, xyyman is talking about Hgs not alleles.
His concept is that white Europeans are depigmenated Africans, not Central Asians and their ancestors lived in Africa less than 5,000 years ago so that's the "we" you are a part of.
To say that that chart is stupid means you are stupid
![[Big Grin]](biggrin.gif)
quote:The link to the peer review is http://file.scirp.org/Html/2-1590439_51973.htm
Originally posted by zarahan- aka Enrique Cardova:
CLyde says:
Of course Klyosov would not have published a paper that shows he is a novice when it comes to population genetics, and the reality the first amh were Black Africans--not whites, as he claims in his numerous misleading papers.
We have discussed other papers by Klyosov on this forum , see: http://www.egyptsearch.com/forums/ultimatebb.cgi?ubb=get_topic;f=15;t=007044;p=1
quote:show me this "we", people talking about alleles in this thread, show me the quote fool
Originally posted by Troll Patrol # Ish Gebor:
LOL at the above. Low level thinking. LOL We are talking about deep clades of alleles. You come here with stupid charts. SMH
quote:LOL. You are very confused. The paper was written by Klysolov. hahahaha
Originally posted by Quetzalcoatl:
quote:The link to the peer review is http://file.scirp.org/Html/2-1590439_51973.htm
Originally posted by zarahan- aka Enrique Cardova:
CLyde says:
Of course Klyosov would not have published a paper that shows he is a novice when it comes to population genetics, and the reality the first amh were Black Africans--not whites, as he claims in his numerous misleading papers.
We have discussed other papers by Klyosov on this forum , see: http://www.egyptsearch.com/forums/ultimatebb.cgi?ubb=get_topic;f=15;t=007044;p=1
The link that is dead is to Clyde Winters 2011 paper because that journal has gone out of business.
All of you are barking up the wrong tree. The peer review IS NOT BY KLYSOLOV. The view deals with Winters's misquoting sources and not presenting evidence for his claims.
quote:Predictably, you have flooded the discussion group with irrelevant threads of spam (Dravidians, the plot of INdutvas, etc.) as a diversion from answering the real criticism in the peer review of your article
Originally posted by Clyde Winters:
This paper as I said earlier does not falsify anything. He was able to dispute the research that shows the first Europeans were not pale,he could not dispute the fact that neither Loschbour nor La Braña carries the skin-lightening allele in SLC24A5…” (Lazaridis et al., 2014), nor was he able to show that the Neanderthal were pale-skinned. Moreover, he did not cite any articles that dispute Boule & Vallois, 1957 research that the Khoisan were the first Europeans.
The rest of the article is his discussion of Africans not being the first anatomically modern humans (amh). This view is not accepted by any modern researchers.Also, I did not mention Indo-Europeans in my paper.
Given the evidence his paper is nothing more than noise, saying nothing relevant. To falsify an article you have to present abundant counter evidence. Any serious reading of this paper will show that the claims in the paper are based on personal opinion not factual and reliable research. he ask the reader to take his word that what he wrote is accurate, in reality it is baseless, and mere ramblings of a man who is living in the past, afraid to admit that the white supremacist ideas he was taught in the past about ancient history are myths.
quote:i.e. that if this paper had had a peer review before publication, as is the usual case, it would not have been published at all.
The cited paper (Winters, 2014) has avoided, unfortunately, an approval by the Editor-in-Chief, and slipped through to the Journal.
quote:Why don't you deal with this rather than floods of spam?
Here is a typical example of the author’s style and reliability of quotations. In his preceding paper, entitled “Possible African origin of Y-chromosome R1-M173” (Winters, 2011), the author writes: “The Khoisan also carry RM343 (R1b)… (Naidoo et al., 2010) the archaeological and linguistic data indicate the successful colonization of Asia by Sub-Saharan Africans from Nubia 5 -4 kya”. If the reader looks up at the Naidoo et al. paper, it shows that the great majority of R1b-M343 haplotypes were found among South African Whites (81 out of 157), while Khoe-san contain three R1b-M343 haplotypes out of 183. Mr. Winters did not even mention such a discrepancy, which brings a different angle at the data.
Here is another example. In the same paper (Winters, 2011) he writes on “widespread distribution of R1*- M173 in Africa, that ranges between 7% - 95% and averages 39% (Coia et al., 2005) ”. He repeats the same “quotation” in yet another paper (Winters, 2010) ―“The frequency of Y-chromosome R1*-M173 in Africa range between 7% - 95% and averages 39.5% (Coia et al., 2005)”. However, in the referenced paper Coia et al. (2005) describe the frequency of R1*-M173 only in Cameroon, “with the highest frequency in North Cameroon (from 6.7% among the Tali to 95.2% among the Uldeme”. Mr. Winters did not mention that the figures were related not to “widespread distribution in Africa”, but specifically to North Cameroon, which is known since at least 2002(Cruciani et al., 2002, 2010) . In the same manner Mr. Winters “quote” data by Berniell-Lee et al. (2009), who had reported that 5.2% of R1b1* were identified in Cameroon and neighboring Gabon, and “quote” it as follows “Haplogroup R1b1* is found in Africa…Berniell-Lee et al. (2005) found in their study that 5.2% carried Rb1*” (spelling by Winters, 2011). As one sees again, Cameroon and Gabon were not mentioned, only “found in Africa 5.2%…”.
Here is yet another example. According to Winters (2011) , “The bearers of R1b1* among the Pygmy populations ranged from 1% - 25% (Berniell-Lee et al., 2009) ”. In fact, Figure 1 in the Berniell-Lee et al. paper shows only two individuals having R1b1*, one from Baka tribe (out of 33 tested) in Gabon, and one from Bakola tribe (out of 22 tested) in Cameroon. So much for 1% - 25%. The list of misquotations can go on. That was a basis for the “African origin of R1-M173”.
quote:More attempts to distract attention from the fact that you misquote papers all the time and you got caught in something that got published and will remain and not disappear as blogs and websites often do.
Originally posted by Clyde Winters:
quote:LOL. You are very confused. The paper was written by Klysolov. hahahaha
Originally posted by Quetzalcoatl:
quote:The link to the peer review is http://file.scirp.org/Html/2-1590439_51973.htm
Originally posted by zarahan- aka Enrique Cardova:
CLyde says:
Of course Klyosov would not have published a paper that shows he is a novice when it comes to population genetics, and the reality the first amh were Black Africans--not whites, as he claims in his numerous misleading papers.
We have discussed other papers by Klyosov on this forum , see: http://www.egyptsearch.com/forums/ultimatebb.cgi?ubb=get_topic;f=15;t=007044;p=1
The link that is dead is to Clyde Winters 2011 paper because that journal has gone out of business.
All of you are barking up the wrong tree. The peer review IS NOT BY KLYSOLOV. The view deals with Winters's misquoting sources and not presenting evidence for his claims.
.
quote:LOL. I know he is the person who wrote the piece because he is listed as the author.hahaha
Originally posted by Quetzalcoatl:
quote:More attempts to distract attention from the fact that you misquote papers all the time and you got caught in something that got published and will remain and not disappear as blogs and websites often do.
Originally posted by Clyde Winters:
quote:LOL. You are very confused. The paper was written by Klysolov. hahahaha
Originally posted by Quetzalcoatl:
quote:The link to the peer review is http://file.scirp.org/Html/2-1590439_51973.htm
Originally posted by zarahan- aka Enrique Cardova:
CLyde says:
Of course Klyosov would not have published a paper that shows he is a novice when it comes to population genetics, and the reality the first amh were Black Africans--not whites, as he claims in his numerous misleading papers.
We have discussed other papers by Klyosov on this forum , see: http://www.egyptsearch.com/forums/ultimatebb.cgi?ubb=get_topic;f=15;t=007044;p=1
The link that is dead is to Clyde Winters 2011 paper because that journal has gone out of business.
All of you are barking up the wrong tree. The peer review IS NOT BY KLYSOLOV. The view deals with Winters's misquoting sources and not presenting evidence for his claims.
.
Exactly how do you know, other than some conspiracy theory, that Klysonov wrote the review? and how does that alter the fact that you misquoted?
quote:The Berniell-Lee article, does say 5% instead of 1-25%.
Originally posted by Quetzalcoatl:
quote:Predictably, you have flooded the discussion group with irrelevant threads of spam (Dravidians, the plot of INdutvas, etc.) as a diversion from answering the real criticism in the peer review of your article
Originally posted by Clyde Winters:
This paper as I said earlier does not falsify anything. He was able to dispute the research that shows the first Europeans were not pale,he could not dispute the fact that neither Loschbour nor La Braña carries the skin-lightening allele in SLC24A5…” (Lazaridis et al., 2014), nor was he able to show that the Neanderthal were pale-skinned. Moreover, he did not cite any articles that dispute Boule & Vallois, 1957 research that the Khoisan were the first Europeans.
The rest of the article is his discussion of Africans not being the first anatomically modern humans (amh). This view is not accepted by any modern researchers.Also, I did not mention Indo-Europeans in my paper.
Given the evidence his paper is nothing more than noise, saying nothing relevant. To falsify an article you have to present abundant counter evidence. Any serious reading of this paper will show that the claims in the paper are based on personal opinion not factual and reliable research. he ask the reader to take his word that what he wrote is accurate, in reality it is baseless, and mere ramblings of a man who is living in the past, afraid to admit that the white supremacist ideas he was taught in the past about ancient history are myths.
1) The reviewer is NOT Klysonov. You don't know anything about the reviewer so your ad hominems about him or her are wastes space.
2) In order to falsify something the first requirement is that there BE something to falsify. It is YOUR job to put together evidence to justify your claim. What the reviewer is pointing out is that you have not presented anything to falsify because your "data" consists of misquoting papers.
3) Apparently you know nothing about peer review. The job is NOT to "disprove the paper being reviewed. It is to see if the paper presents sufficient evidence supporting the claims, whether the results are something new, whether it is appropriate for the journal in question, whether it is important enough to be published, whether the documentation, footnoting, and coverage of the literature is sufficient. What the reviewer is pointing out is that to quote the reviewquote:i.e. that if this paper had had a peer review before publication, as is the usual case, it would not have been published at all.
The cited paper (Winters, 2014) has avoided, unfortunately, an approval by the Editor-in-Chief, and slipped through to the Journal.
4) Here there and everywhere you have claimed that R-M173 is "widespread in Africa" but this is not true and you have not presented evidence that it is. You have cited papers about a narrow area of Africa that has R-V88 and pretended that these papers supported your claim of a wide distribution. From the review:quote:Why don't you deal with this rather than floods of spam?
Here is a typical example of the author’s style and reliability of quotations. In his preceding paper, entitled “Possible African origin of Y-chromosome R1-M173” (Winters, 2011), the author writes: “The Khoisan also carry RM343 (R1b)… (Naidoo et al., 2010) the archaeological and linguistic data indicate the successful colonization of Asia by Sub-Saharan Africans from Nubia 5 -4 kya”. If the reader looks up at the Naidoo et al. paper, it shows that the great majority of R1b-M343 haplotypes were found among South African Whites (81 out of 157), while Khoe-san contain three R1b-M343 haplotypes out of 183. Mr. Winters did not even mention such a discrepancy, which brings a different angle at the data.
Here is another example. In the same paper (Winters, 2011) he writes on “widespread distribution of R1*- M173 in Africa, that ranges between 7% - 95% and averages 39% (Coia et al., 2005) ”. He repeats the same “quotation” in yet another paper (Winters, 2010) ―“The frequency of Y-chromosome R1*-M173 in Africa range between 7% - 95% and averages 39.5% (Coia et al., 2005)”. However, in the referenced paper Coia et al. (2005) describe the frequency of R1*-M173 only in Cameroon, “with the highest frequency in North Cameroon (from 6.7% among the Tali to 95.2% among the Uldeme”. Mr. Winters did not mention that the figures were related not to “widespread distribution in Africa”, but specifically to North Cameroon, which is known since at least 2002(Cruciani et al., 2002, 2010) . In the same manner Mr. Winters “quote” data by Berniell-Lee et al. (2009), who had reported that 5.2% of R1b1* were identified in Cameroon and neighboring Gabon, and “quote” it as follows “Haplogroup R1b1* is found in Africa…Berniell-Lee et al. (2005) found in their study that 5.2% carried Rb1*” (spelling by Winters, 2011). As one sees again, Cameroon and Gabon were not mentioned, only “found in Africa 5.2%…”.
Here is yet another example. According to Winters (2011) , “The bearers of R1b1* among the Pygmy populations ranged from 1% - 25% (Berniell-Lee et al., 2009) ”. In fact, Figure 1 in the Berniell-Lee et al. paper shows only two individuals having R1b1*, one from Baka tribe (out of 33 tested) in Gabon, and one from Bakola tribe (out of 22 tested) in Cameroon. So much for 1% - 25%. The list of misquotations can go on. That was a basis for the “African origin of R1-M173”.
quote:As you can see I did admit that only 5.2% of the pygmy carried R1.
Y-chromosome R1 is found throghout Africa. The pristine
form of R1-M173 is only found in Africa (Coia et al,
2005; Cruciani et al, 2002, 2010). The age of ychromosome
R is 27ky. Most researchers believe that
R(M173) is 18.5 ky.There is a great diversity of the
macrohaplogroup R in Africa (See Figure 1). Ychromosome
R is characterized by M207/V45. The V45
mutation is found among African populations ( Cruciani et
al ,2010). ISOGG 2010 Y-DNA haplogroup tree makes it
clear that V45 is phylogenetically equivalent to M207.The
most common R haplogroup in Africa is R1 (M173). The
predominant haplogroup is R1b (Berniell-Lee et al,2009;
Coia et al, 2005; Winters, 2010b; Wood et al, 2009).
Cruciani et al (2010) discovered new R1b mutations
including V7, V8, V45, V69, and V88. Geography appears
to play a significant role in the distribution of haplogroup
R in Africa. Cruciani et al (2010) has renamed the R*-
M173 (R P-25) in Africa V88. The TMRCA of V88 was
9200-5600 kya (Cruciani et al, 2010).
Y-chromosome V88 (R1b1a) has its highest frequency
among Chadic speakers, while the carriers of V88 among
Niger-Congo speakers (predominately Bantu people)
range between 2-66% ( Cruciani et al, 2010; Bernielle-Lee
et al, 2009). Haplogroup V88 includes the mutations M18,
V35 and V7. Cruciani et al (2010) revealed that R-V88 is
also carried by Eurasians including the distinctive
mutations M18, V35 and V7.
R1b1-P25 is found in Western Eurasia. Haplogroup
R1b1* is found in Africa at various frequencies. BerniellLee
et al (2009) found in their study that 5.2% carried
Rb1*. The frequency of R1b1* among the Bantu ranged
from 2-20. The bearers of R1b1* among the Pygmy
populations ranged from 1-25% (Berniell-Lee et al, 2009).
The frequency of R1b1 among Guinea-Bissau populations
was 12% (Carvalho et al,2010).
quote:As a result, I did not attempt to decieve anyone about the frequency of R1 in Africa as the author implies.
Around 0.1 of Sub Saharan Africans carry R1b1b2. Wood
et al (2009) found that Khoisan (2.2%) and Niger-Congo
(0.4%) speakers carried the R-M269 y-chromosome.
The Khoisan also carry RM343 (R1b) and M 198 (R1a1)
(Naidoo et al., 2010) the archaeological and linguistic data
indicate the successful colonization of Asia by SubSaharan
Africans from Nubia 5-4kya (Winters, 2007,2008,
2010c). The archaeological evidence makes it clear that
around 4kya intercultural style artifacts connected Africa
and Eurasia (Winters, 2007,2010c).

quote:
Originally posted by Clyde Winters:
Exactly how do you know, other than some conspiracy theory, that Klysonov wrote the review? and how does that alter the fact that you misquoted?
quote:You are right. I went back to see how the paper was supposed to be cited and Klysonov is listed as the author.
LOL. I know he is the person who wrote the piece because he is listed as the author.hahaha
quote:‘‘Out of Africa’’ haplogroups.
Originally posted by the lioness,:
quote:show me this "we", people talking about alleles in this thread, show me the quote fool
Originally posted by Troll Patrol # Ish Gebor:
LOL at the above. Low level thinking. LOL We are talking about deep clades of alleles. You come here with stupid charts. SMH
![[Big Grin]](biggrin.gif)
quote:R is not found throughout Africa
Originally posted by Clyde Winters:
[QB] Klyosov in criticizing my article exagerates what I wrote. He implies that I was trying to decieve the readers about the frequency of R1 in Africa. This is false.
I specifically stated the frequency of R1 among African populations throughout my 2011 paper.
[QUOTE]
Y-chromosome R1 is found throughout Africa. The pristine
form of R1-M173 is only found in Africa (Coia et al,
2005; Cruciani et al, 2002, 2010).
quote:
Y-Chromosome Variation Among Sudanese:Restricted Gene Flow, Concordance With Language, Geography, and History
Hisham Y. Hassan 2008
Haplogroup R-M173 appears to be the most frequent haplogroup in Fulani,
The Fulani, who possess the lowest population size inthis study, have an interesting genetic structure, effectively consisting of two haplogroups or founding line-ages. One of the lineages is R-M173 (53.8%), and its sheer frequency suggests either a recent migration of this group to Africa and/or a restricted gene flow due to linguistic or cultural barriers. The high frequency of sub-clade E-V22, which is believed to be northeast African (Cruciani et al., 2007) and haplogroup R-M173, suggests an amalgamation of two populations/cultures that took place sometime in the past in eastern or central Africa.This is also evident from the frequency of the ‘‘T’’ allele of the lactase persistence gene that is uniquely present in considerable frequencies among the Fulani (Mulcareet al., 2004). Interestingly, Fulani language is classified in the Niger-Congo family of languages, which is more prevalent in West Africa and among Bantu speakers, yet their Y-chromosomes show very little evidence of West African genetic affiliation.

quote:
Originally posted by Clyde Winters:
my 2011 paper.
quote:
Cruciani et al (2010) has renamed the R*-
M173 (R P-25) in Africa V88. The TMRCA of V88 was
9200-5600 kya (Cruciani et al, 2010).

quote:
The phylogeographic profile of R1*-M173 supports this ancient migration of Kushites from Africa to Eurasia as suggested by the Classical writers. This expansion of Kushites into Eurasia probably took place over 4 kya.
--Clyde Winters
The Kushite Spread of Haplogroup R1*-M173 from Africa to Eurasia


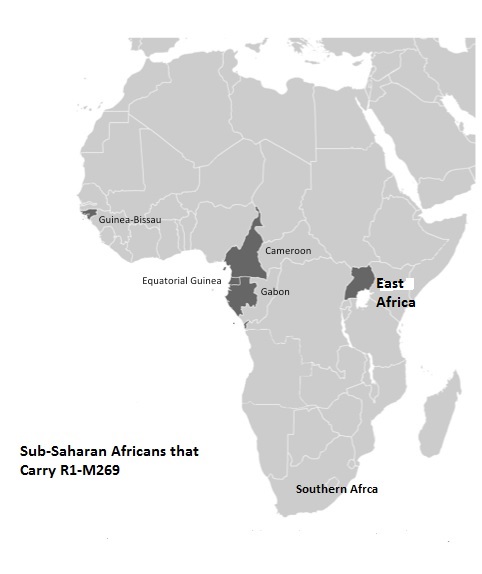




quote:
Originally posted by the lioness,:
Clyde, on what continent did the first clade of Haplogroup R called
R-M207 originate ?

quote:that is incorrect
Originally posted by Clyde Winters:
R-M207 is the same as V45.
quote:^^^ here they show V5 as an allele of 207
R M207/UTY2, P224, P227, P229, P232, P280,
P285, S4, S8, S9, V45
quote:^^^ added SNPs > reference Cruciani et al (2010) >>>
Corrections/Additions made since 1 January 2010:
Added the following SNPs L165, L193, L196, L226, P89.2, V7, V8, V35, V45, V69, V88 and Cruciani et al (2010) on 12 January 2010
quote:^^^ this is the sole and primary source about V45 (discovered in 2010)

We resequenced about 0.15 Mb of the MSY for each of the four R1b subjects and found six new mutations (V7, V8, V35, V45, V69, and V88). The V45 mutation is phylogenetically equivalent to M173. Among the other five mutations, V88 defines a new monophyletic clade (R-V88 or R1b1a), which includes haplogroups R-M18 (R1b1a1, formerly R1b1a), R-V8 (R1b1a2), R-V35 (R1b1a3, further subdivided by the V7 mutation to R1b1a3* and R1b1a3a), and R-V69 (R1b1a4) (Figure 1).
-- Human Y chromosome haplogroup R-V88: a paternal genetic record of early mid Holocene trans-Saharan connections and the spread of Chadic languages
Fulvio Cruciani, 2010
quote:LOL. They can change whatever they wish. I still believe it originated in Africa.
Originally posted by the lioness,:
quote:that is incorrect
Originally posted by Clyde Winters:
R-M207 is the same as V45.
ISOGG made an error or correction on this
http://www.isogg.org/tree/ISOGG_HapgrpR10.html
quote:^^^ here they show V5 as an allele of 207
R M207/UTY2, P224, P227, P229, P232, P280,
P285, S4, S8, S9, V45
However on the same page:
quote:^^^ added SNPs > reference Cruciani et al (2010) >>>
Corrections/Additions made since 1 January 2010:
Added the following SNPs L165, L193, L196, L226, P89.2, V7, V8, V35, V45, V69, V88 and Cruciani et al (2010) on 12 January 2010
quote:^^^ this is the sole and primary source about V45

We resequenced about 0.15 Mb of the MSY for each of the four R1b subjects and found six new mutations (V7, V8, V35, V45, V69, and V88). The V45 mutation is phylogenetically equivalent to M173. Among the other five mutations, V88 defines a new monophyletic clade (R-V88 or R1b1a), which includes haplogroups R-M18 (R1b1a1, formerly R1b1a), R-V8 (R1b1a2), R-V35 (R1b1a3, further subdivided by the V7 mutation to R1b1a3* and R1b1a3a), and R-V69 (R1b1a4) (Figure 1).
-- Human Y chromosome haplogroup R-V88: a paternal genetic record of early mid Holocene trans-Saharan connections and the spread of Chadic languages
Fulvio Cruciani, 2010
The chart above is for R1
However R-M207 is ancestor to R1
That's the paragroup, the R* root before R1 came into existence
quote:So that means the people of Wales are 92.3% African paternally
Originally posted by Clyde Winters:
The fact remains that R-M269, is found among Sub-Saharan Africans from West, to East and Southern Africa. This supports my contention that this haplogroup is widespread in Africa.
.
.
quote:LOL.I am not joking, your statement on the otherhand about V45, is further support to the view that Europeans love to lie. I can laugh at the statement about V45, now being M173, when it was M207 in 2011. Geneticist knew what chracterized M173, and M207 in 2011. So I have to believe that they changed the nomenclature of V45, because it supported an African origin for haplogroup R.
Originally posted by the lioness,:
quote:So that means the people of Wales are 92.3% African paternally
Originally posted by Clyde Winters:
The fact remains that R-M269, is found among Sub-Saharan Africans from West, to East and Southern Africa. This supports my contention that this haplogroup is widespread in Africa.
.
.
rather than Central Asian because M269 originates in Africa, correct ?
Additionally
M269
Basques, Spain 87.1
Ireland 85.4
France 80.5
England 62.0
etc, etc all over Europe, high frequencies
So Clyde, you would agree with xyyman, modern Europeans are depigmented Africans rather than Central Asians right?
Their Y DNA is primarily R and R is an African haplogroup
Don't LOL me, this is pure logic stemming directly from what you are saying
The simple fact is that if what you are claiming is true, that R1 is an African haplogroup and modern Europeans primarily carry R1 as their Y-DNA then there is no escaping the fact that they are primarily African paternally
Either that is bizarre but true
or your claim is not true, so don't try to play like I'm joking
quote:Hum, let's see?
Originally posted by the lioness,:
quote:
Originally posted by Clyde Winters:
my 2011 paper.
quote:
Cruciani et al (2010) has renamed the R*-
M173 (R P-25) in Africa V88. The TMRCA of V88 was
9200-5600 kya (Cruciani et al, 2010).
M173 (R P-25) renamed V88 as per alleles specific to Africa.
quote:
The phylogeographic profile of R1*-M173 supports this ancient migration of Kushites from Africa to Eurasia as suggested by the Classical writers. This expansion of Kushites into Eurasia probably took place over 4 kya.
--Clyde Winters
The Kushite Spread of Haplogroup R1*-M173 from Africa to Eurasia
.
Clyde, on what continent did Haplgroup R originate ?
.
![[Big Grin]](biggrin.gif)
quote:DE was found in Africa, amongst Africans. CT is at the root in Africa out.
Originally posted by the lioness,:
quote:So that means the people of Wales are 92.3% African paternally
Originally posted by Clyde Winters:
The fact remains that R-M269, is found among Sub-Saharan Africans from West, to East and Southern Africa. This supports my contention that this haplogroup is widespread in Africa.
.
.
rather than Central Asian because M269 originates in Africa, correct ?
Additionally
M269
Basques, Spain 87.1
Ireland 85.4
France 80.5
England 62.0
etc, etc all over Europe, high frequencies
So Clyde, you would agree with xyyman, modern Europeans are depigmented Africans rather than Central Asians right?
Their Y DNA is primarily R and R is an African haplogroup
Don't LOL me, this is pure logic stemming directly from what you are saying
The simple fact is that if what you are claiming is true, that R1 is an African haplogroup and modern Europeans primarily carry R1 as their Y-DNA then there is no escaping the fact that they are primarily African paternally
Either that is bizarre but true
or your claim is not true, so don't try to play like I'm joking
quote:There you go again!!! Exactly what the review of your paper was critizing. You are misquoting your sources to create a false impression supporting your idiosyncratic view.
Originally posted by Clyde Winters:
The fact remains that R-M269, is found among Sub-Saharan Africans from West, to East and Southern Africa and include Niger-Congo speakers, Pgymies, and Khoisan . This supports my contention that this haplogroup is widespread in Africa.
quote:[/QUOTE]
p.327 Altogether, the proportion of recent Eurasian admixture found in our sample is approximately 15% (haplogroups E1b1b1b-M81, G-M201, N1c-Tat, R1b1b2-M269 and two chromosomes belonging to E1b1b1a-M78—see criteria below), which is easily explained by the well-reported European arrivals to this territory within the last five centuries.
328 A phylogenetic network of R1b1 lineages based on 10 Y-STR
haplotypes was constructed with samples from Cameroon and
Gabon5 and from the present study (Figure 2a). A clear separation of the R1b1b2-M269 samples was observed; they clustered with the European modal haplotype, supporting their European ancestry.
quote:You never acknowledge error or learn from them. In 2011, you were shown an error affecting your 2010 M-173 paper, which was also criticized in the peer review.
A group of chromosomes of potential interest to past trans-Saharan connections is the paragroup R1b1* (R-P25*). Cruciani et al18 found this paragroup (at that time defined as haplogroup 117, or R-M173*(xSRY10831, M18, M73, M269)) to be present at high frequencies (up to 95%) in populations from northern Cameroon. The same paragroup was only rarely observed in other sub-Saharan African regions, and not observed at all in western Eurasia.18 Subsequent studies dealing with the MSY diversity in Africa have confirmed the presence of R-P25*(xM269)[BOM excluding M269) in northern Cameroon at high frequencies 23and, at lower frequencies (mean 5%, range 0–20%), of R-P25* immediately south of Cameroon, in several populations from Gabon.
quote:In the Cruciani paper quoted above R-P25*(xM269) means that all the clades of R-P25* except the M269, i.e. R-V88 are found in Cameroon.
http://www.egyptsearch.com/forums/ultimatebb.cgi?ubb=get_topic;f=15;t=003788 Originally posted by Clyde Winters:
• Figure 2: Distribution of R1b among African Populations
Mandekan………..…………..…100%
Mossi………….……………….100%
Rimaiba………….…………….100%
Fulbe(Burkina)…….…………..91.0%
Fulbe(Niger)…………………..85.7%
Fulbe(Nigeria)…………………80.0%
Fulbe(Cameroon)……..………88.9%
Bamileke……………………….100%
Ewondo………………………….96.7
Biaka (Pygmies)………………..100%
Mbuti(Pygmies)……..………...100%
Twa(Pygmies)………………….100%
This figure is based on Cruciani et al (2010).
Posted by alTakruri (Member # 10195) on 20 February, 2011 12:15 AM:
Why won't you understand that Y(xR1b)* means every haplogroup except R1b?
This is basic population genetics terminology:
Y = the entire set of MSY chromosome haplogroups
x = except
R1b = R1b including its downstream subclades.
All the other columns in Table 1 are telling you the percise R1b of the subjects.
For example row 1 is of 55 sampled Moroccan Arabs where 1.8% have R1b1b2
but no other R1b haplogroups while the remaing 98.2% have haplogroups that
are neither R1b nor any of its subclades at all. There's nothing to debate here.
I know what table the data comes from as seen by the haplogroups and their
frequencies provided in my earlier post. Check the figures yourself, you'll see.
%%%%%%
[
Posted by Clyde Winters (Member # 10129) on 20 February, 2011 01:33 AM:
quote:
Originally posted by alTakruri:
Why won't you understand that Y(xR1b)* means every haplogroup except R1b?
This is basic population genetics terminology:
Y = the entire set of MSY chromosome haplogroups
x = except
R1b = R1b including its downstream subclades.
All the other columns in Table 1 are telling you the percise R1b of the subjects.
For example row 1 is of 55 sampled Moroccan Arabs where 1.8% have R1b1b2
but no other R1b haplogroups while the remaing 98.2% have haplogroups that
are neither R1b nor any of its subclades at all. There's nothing to debate here.
I know what table the data comes from as seen by the haplogroups and their
frequencies provided in my earlier post. Check the figures yourself, you'll see.
BTW Congratulations on your two peer review published articles in Curr Res J Biol Sci.
quote:
Originally posted by Clyde Winters:
These figures were taken from this Table
quote:
Originally posted by alTakruri:
Mandekan, Mossi, Rimaiba, Bamileke, Biaka, Mbuti, and
Twa have no R1b MSY chromosomes in the reported samples.
Fulbe(Burkina) have no R1b
Fulbe(Niger) have 14.3% R1b as R-V88*
Fulbe(Nigeria) have no R1b
Fulbe(Cameroon) have 11.1% R1b as 5.6% R-V88* plus 5.6% R-V69
Ewondo have 3.3% R1b as R-V88*
Above are the correct readings
of Table 1 in Crucuani (2010).
Outside of Africa R-V88 showed up in
2 out of 1173 Italians (0.2%), one had subclade R-V35 the other R-V7
1 out of 141 Corsicans (0.7%), he had subclade R-M18
1 out of 510 Balkans (0.2%) and
1 out of 328 Western Asians (0.3%).
5 out of 2152 amounts to 2/10ths of 1%.
I read the table wrong. I did not know that x=except. Thanks for making it clear.I would have been embarassed if I had made these statements in an article.
I will admit I am still a novice in this area. But I am still trying to project a different interpretation of the data using archaeogenetics.
Thanks again for the heads up.
quote:Now to your figures:
According to the phylogeography of macro-haplogroup K-M9
(which contains haplogroup R1b), an ancient Asia-to-Africa back
migration has been hypothesized to explain the puzzling presence of R-P25* in sub-Saharan Africa.18 This hypothesis is strongly supported by the present data. In the revised Y chromosome phylogeny, there are 119 lineages in the macro-haplogroup K-M9 (which includes haplogroups K1-K4 and L to T).31 Of these lineages, only two have been observed in sub-Saharan Africa at appreciable frequencies: T-M7018,41,42 and R-V88 (this study). Both haplogroups have also been observed in Europe and western Asia (Refs 42,43 and this study). If the presence of R1b chromosomes in Africa was not because of a back migration, we would have to assume that all the mutations that connect M9 with V88 in the MSY phylogeny (450 mutations) originated in Africa. Under this scenario, we should assume that all the K-M9 lineages that are now found outside sub-Saharan Africa have survived extinction, whereas those which should have accumulated
in Africa are now extinct (with the exception of T-M70 and
R-V88) and this is an unlikely scenario.
quote:In a later post I show that you are misinterpreting/misquoting these sources. R-M269 is NOT African but European and the only place with significant R1 is Cameroon where the haplotype is R-V88.
Originally posted by Clyde Winters:
Klyosov in criticizing my article exagerates what I wrote. He implies that I was trying to decieve the readers about the frequency of R1 in Africa. This is false.
I specifically stated the frequency of R1 among African populations throughout my 2011 paper.
quote:As you can see I did admit that only 5.2% of the pygmy carried R1.
Y-chromosome R1 is found throghout Africa. The pristine
form of R1-M173 is only found in Africa (Coia et al,
2005; Cruciani et al, 2002, 2010). The age of ychromosome
R is 27ky. Most researchers believe that
R(M173) is 18.5 ky.There is a great diversity of the
macrohaplogroup R in Africa (See Figure 1). Ychromosome
R is characterized by M207/V45. The V45
mutation is found among African populations ( Cruciani et
al ,2010). ISOGG 2010 Y-DNA haplogroup tree makes it
clear that V45 is phylogenetically equivalent to M207.The
most common R haplogroup in Africa is R1 (M173). The
predominant haplogroup is R1b (Berniell-Lee et al,2009;
Coia et al, 2005; Winters, 2010b; Wood et al, 2009).
Cruciani et al (2010) discovered new R1b mutations
including V7, V8, V45, V69, and V88. Geography appears
to play a significant role in the distribution of haplogroup
R in Africa. Cruciani et al (2010) has renamed the R*-
M173 (R P-25) in Africa V88. The TMRCA of V88 was
9200-5600 kya (Cruciani et al, 2010).
Y-chromosome V88 (R1b1a) has its highest frequency
among Chadic speakers, while the carriers of V88 among
Niger-Congo speakers (predominately Bantu people)
range between 2-66% ( Cruciani et al, 2010; Bernielle-Lee
et al, 2009). Haplogroup V88 includes the mutations M18,
V35 and V7. Cruciani et al (2010) revealed that R-V88 is
also carried by Eurasians including the distinctive
mutations M18, V35 and V7.
R1b1-P25 is found in Western Eurasia. Haplogroup
R1b1* is found in Africa at various frequencies. BerniellLee
et al (2009) found in their study that 5.2% carried
Rb1*. The frequency of R1b1* among the Bantu ranged
from 2-20. The bearers of R1b1* among the Pygmy
populations ranged from 1-25% (Berniell-Lee et al, 2009).
The frequency of R1b1 among Guinea-Bissau populations
was 12% (Carvalho et al,2010).
In relation to R-M269 in Africa I wrote:
quote:As a result, I did not attempt to decieve anyone about the frequency of R1 in Africa as the author implies.
Around 0.1 of Sub Saharan Africans carry R1b1b2. Wood
et al (2009) found that Khoisan (2.2%) and Niger-Congo
(0.4%) speakers carried the R-M269 y-chromosome.
The Khoisan also carry RM343 (R1b) and M 198 (R1a1)
(Naidoo et al., 2010) the archaeological and linguistic data
indicate the successful colonization of Asia by SubSaharan
Africans from Nubia 5-4kya (Winters, 2007,2008,
2010c). The archaeological evidence makes it clear that
around 4kya intercultural style artifacts connected Africa
and Eurasia (Winters, 2007,2010c).
In fact recent research on y-haplogroups in Africa suggest that R1-M269 is also widespread in Africa.
Above is a figure from Gonzalez et al. The Gonzalez et al article found that 10 out of 19 subjects in the study carried R1b1-P25 or M269. This is highly significant because it indicates that 53% of the R1 carriers in this study were M269, this finding is further proof of the widespread nature of this so-called Eurasian genes in Africa among populations that have not mated with Europeans.
quote:However your information is obsolete due to recent analysis of Siberian DNA
Originally posted by Clyde Winters:
M207, like the rest of the R macrohaplogroup originated in Africa.
quote:Clyde if V88 is as young as 9200-5600 kya why even bother mentioning it?
Originally posted by Clyde Winters:
, I argue that the P clade originated in Africa because 1) the age of R-V88 and 2) the widespread nature of R1 in Africa. Researchers have found that the TMRCA of V88 was 9200-5600 kya (Cruciani et al, 2010).
quote:
Originally posted by the lioness,:
quote:However your information is obsolete due to recent analysis of Siberian DNA
Originally posted by Clyde Winters:
M207, like the rest of the R macrohaplogroup originated in Africa.
Haplogroup R* was found in the remains of a Palaeolithic boy MA-1 (Mal'ta) near Lake Baikal in Siberia, dating to 24,000 years ago.
quote:Lol- this kind of bigotry is getting hilarious.
Originally posted by Quetzalcoatl:
quote:Use of Y Chromosome and Mitochondrial DNA Population Structure in Tracing Human Migrations
"haplogroup CF and DE molecular ancestors first evolved inside Africa and subsequently contributed as Y chromosome founders to pioneering migrations that successfully colonized Asia. While not proof, the DE and CF bifurcation (Figure 8d ) is consistent with independent colonization impulses possibly occurring in a short time interval."
quote:
Originally posted by Clyde Winters:

quote:You would have to look at each reference that Clyde has cited already if you haven't done it already.
Originally posted by Quetzalcoatl:
In a later post I show that you are misinterpreting/misquoting these sources. R-M269 is NOT African but European and the only place with significant R1 is Cameroon where the haplotype is R-V88.
quote:
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3573200/
The genetic landscape of Equatorial Guinea and the origin and migration routes of the Y chromosome haplogroup R-V88
Miguel González,1
Altogether, the proportion of recent Eurasian admixture found in our sample is approximately 15% (haplogroups E1b1b1b-M81, G-M201, N1c-Tat, R1b1b2-M269 and two chromosomes belonging to E1b1b1a-M78—see criteria below), which is easily explained by the well-reported European arrivals to this territory within the last five centuries.
A phylogenetic network of R1b1 lineages based on 10 Y-STR haplotypes was constructed with samples from Cameroon and Gabon5 and from the present study (Figure 2a). A clear separation of the R1b1b2-M269 samples was observed; they
clustered with the European modal haplotype, supporting their European ancestry. The non-consensus alleles observed in both studies do not show a well-defined separation from the samples with consensus alleles. However, a cluster containing haplotypes with both consensus and non-consensus alleles (representing two different lineages) and another exclusively with consensus alleles could be indicative of at least three different lineages within the R1b1-P25( × M269) haplogroup. Nonetheless, it is important to note that the data from Berniell-Lee et al5 were published before the recently discovered mutations within P25.7 Cruciani et al7 did not find any intermediate variant alleles at the locus DYS385, which were all found within haplogroup R1b1a-V88 in the present work (with only two chromosomes presenting consensus alleles). This result may indicate the presence of different sub-lineages within this haplogroup that are yet to be discovered.
quote:You must have a new definition of bigotry because Haplogroup R* was found in the remains of a Palaeolithic boy MA-1 (Mal'ta) near Lake Baikal in Siberia,
Originally posted by Troll Patrol # Ish Gebor:
quote:
Originally posted by the lioness,:
[qb]quote:However your information is obsolete due to recent analysis of Siberian DNA
Originally posted by Clyde Winters:
M207, like the rest of the R macrohaplogroup originated in Africa.
Haplogroup R* was found in the remains of a Palaeolithic boy MA-1 (Mal'ta) near Lake Baikal in Siberia, dating to 24,000 years ago.quote:Lol- this kind of bigotry is getting hilarious.
Originally posted by Quetzalcoatl:
quote:
Y-DNA haplogroup R-M207 is believed to have arisen approximately 27,000 years ago in Asia. The two currently defined subclades are R1 and R2.
R1-M173 is estimated to have arisen during the height of the Last Glacial Maximum (LGM), about 18,500 years ago, most likely in southwestern Asia. The two most common descendant clades of R1 are R1a and R1b.
R1a-M420 is believed to have arisen on the Eurasian Steppe or the Indus Valley, and today is most frequently observed in eastern Europe and in western and central Asia. R1a1a1g-M458 is found at frequencies approaching or exceeding 30% in Eastern Europe.
R1b-M343 is believed to have arisen in southwest Asia and today its sublcades are bound in various distributions across Eurasia and Africa.
Paragroup R1b1* and R1b1c-V88 are found most frequently in SW Asia and Africa. The African examples are almost entirely within R1b1c and are associated with the spread of Chadic languages.
R1b1a-P297 is found throughout Eurasia. R1b1a1-M73 is observed most frequently in Asia, with low frequency of observation in Europe. R1b1a2-M269 is observed most frequently in Europe, especially western Europe, but with notable frequency in southwest Asia.
R1b1a2-M269 is estimated to have arisen approximately 4,000 to 8,000 years ago in southwest Asia and to have spread into Europe from there. The Atlantic Modal Haplotype, or AMH, is the most common STR haplotype in R1b1a2a1a-L11/PF6539/S127 and most European R1b1a2 belongs to R1b1a2a1a1-M405/S21/U106 or R1b1a2a1a2-P312/PF6547/S116.
R2-M479 is most often observed in Asia, especially on the Indian sub-continent and in central Asia.
quote:LOL. Please answer these questions:
Originally posted by the lioness,:
Clyde's chart
quote:
Originally posted by Clyde Winters:

quote:You would have to look at each reference that Cloyed has cited already if you haven't done it already.
Originally posted by Quetzalcoatl:
In a later post I show that you are misinterpreting/misquoting these sources. R-M269 is NOT African but European and the only place with significant R1 is Cameroon where the haplotype is R-V88.
On the top line
Berniel Lee 2009
it says Africa 5.2%
Yet in Europe it's about 55%
Clyde's theory fails right there
Additionally there is a much greater diversity of R outside of Africa
I said to Cloyed this would mean modern Europeans are paternally African
But Cloyed went silent on that one. That silence is a fail
He has no answer for it
______________________
Now let's look high percentage of M269 he refers to
in the 2012 Gonzalez article, 53%
I'm not sure where he got the percentage from (Clyde where in the paper or Supplement is that 53% you made it up?)
Equatorial Guinea? the place is tiny
Hap R is very very sparse in Africa
An exceptional region is particular groups in Cameroon Chad who carry V88
Below the quotes pertaining to M269
quote:
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3573200/
The genetic landscape of Equatorial Guinea and the origin and migration routes of the Y chromosome haplogroup R-V88
Miguel González,1
Altogether, the proportion of recent Eurasian admixture found in our sample is approximately 15% (haplogroups E1b1b1b-M81, G-M201, N1c-Tat, R1b1b2-M269 and two chromosomes belonging to E1b1b1a-M78—see criteria below), which is easily explained by the well-reported European arrivals to this territory within the last five centuries.
A phylogenetic network of R1b1 lineages based on 10 Y-STR haplotypes was constructed with samples from Cameroon and Gabon5 and from the present study (Figure 2a). A clear separation of the R1b1b2-M269 samples was observed; they
clustered with the European modal haplotype, supporting their European ancestry. The non-consensus alleles observed in both studies do not show a well-defined separation from the samples with consensus alleles. However, a cluster containing haplotypes with both consensus and non-consensus alleles (representing two different lineages) and another exclusively with consensus alleles could be indicative of at least three different lineages within the R1b1-P25( × M269) haplogroup. Nonetheless, it is important to note that the data from Berniell-Lee et al5 were published before the recently discovered mutations within P25.7 Cruciani et al7 did not find any intermediate variant alleles at the locus DYS385, which were all found within haplogroup R1b1a-V88 in the present work (with only two chromosomes presenting consensus alleles). This result may indicate the presence of different sub-lineages within this haplogroup that are yet to be discovered.

quote:
Originally posted by the lioness,:
quote:You must have a new definition of bigotry because Haplogroup R* was found in the remains of a Palaeolithic boy MA-1 (Mal'ta) near Lake Baikal in Siberia,
Originally posted by Troll Patrol # Ish Gebor:
quote:
Originally posted by the lioness,:
[qb]quote:However your information is obsolete due to recent analysis of Siberian DNA
Originally posted by Clyde Winters:
M207, like the rest of the R macrohaplogroup originated in Africa.
Haplogroup R* was found in the remains of a Palaeolithic boy MA-1 (Mal'ta) near Lake Baikal in Siberia, dating to 24,000 years ago.quote:Lol- this kind of bigotry is getting hilarious.
Originally posted by Quetzalcoatl:
Why woud such a fact be bigotry?
http://www.isogg.org/tree/ISOGG_HapgrpR.html
ISOGG
quote:
Y-DNA haplogroup R-M207 is believed to have arisen approximately 27,000 years ago in Asia. The two currently defined subclades are R1 and R2.
R1-M173 is estimated to have arisen during the height of the Last Glacial Maximum (LGM), about 18,500 years ago, most likely in southwestern Asia. The two most common descendant clades of R1 are R1a and R1b.
R1a-M420 is believed to have arisen on the Eurasian Steppe or the Indus Valley, and today is most frequently observed in eastern Europe and in western and central Asia. R1a1a1g-M458 is found at frequencies approaching or exceeding 30% in Eastern Europe.
R1b-M343 is believed to have arisen in southwest Asia and today its sublcades are bound in various distributions across Eurasia and Africa.
Paragroup R1b1* and R1b1c-V88 are found most frequently in SW Asia and Africa. The African examples are almost entirely within R1b1c and are associated with the spread of Chadic languages.
R1b1a-P297 is found throughout Eurasia. R1b1a1-M73 is observed most frequently in Asia, with low frequency of observation in Europe. R1b1a2-M269 is observed most frequently in Europe, especially western Europe, but with notable frequency in southwest Asia.
R1b1a2-M269 is estimated to have arisen approximately 4,000 to 8,000 years ago in southwest Asia and to have spread into Europe from there. The Atlantic Modal Haplotype, or AMH, is the most common STR haplotype in R1b1a2a1a-L11/PF6539/S127 and most European R1b1a2 belongs to R1b1a2a1a1-M405/S21/U106 or R1b1a2a1a2-P312/PF6547/S116.
R2-M479 is most often observed in Asia, especially on the Indian sub-continent and in central Asia.
quote:LOL. How can I be misquoting Gonzalez et al (2012), the figure is above and you can count the number of M269 subjects found in the study yourself.
Originally posted by Quetzalcoatl:
quote:In a later post I show that you are misinterpreting/misquoting these sources. R-M269 is NOT African but European and the only place with significant R1 is Cameroon where the haplotype is R-V88.
Originally posted by Clyde Winters:
Klyosov in criticizing my article exagerates what I wrote. He implies that I was trying to decieve the readers about the frequency of R1 in Africa. This is false.
I specifically stated the frequency of R1 among African populations throughout my 2011 paper.
quote:As you can see I did admit that only 5.2% of the pygmy carried R1.
Y-chromosome R1 is found throghout Africa. The pristine
form of R1-M173 is only found in Africa (Coia et al,
2005; Cruciani et al, 2002, 2010). The age of ychromosome
R is 27ky. Most researchers believe that
R(M173) is 18.5 ky.There is a great diversity of the
macrohaplogroup R in Africa (See Figure 1). Ychromosome
R is characterized by M207/V45. The V45
mutation is found among African populations ( Cruciani et
al ,2010). ISOGG 2010 Y-DNA haplogroup tree makes it
clear that V45 is phylogenetically equivalent to M207.The
most common R haplogroup in Africa is R1 (M173). The
predominant haplogroup is R1b (Berniell-Lee et al,2009;
Coia et al, 2005; Winters, 2010b; Wood et al, 2009).
Cruciani et al (2010) discovered new R1b mutations
including V7, V8, V45, V69, and V88. Geography appears
to play a significant role in the distribution of haplogroup
R in Africa. Cruciani et al (2010) has renamed the R*-
M173 (R P-25) in Africa V88. The TMRCA of V88 was
9200-5600 kya (Cruciani et al, 2010).
Y-chromosome V88 (R1b1a) has its highest frequency
among Chadic speakers, while the carriers of V88 among
Niger-Congo speakers (predominately Bantu people)
range between 2-66% ( Cruciani et al, 2010; Bernielle-Lee
et al, 2009). Haplogroup V88 includes the mutations M18,
V35 and V7. Cruciani et al (2010) revealed that R-V88 is
also carried by Eurasians including the distinctive
mutations M18, V35 and V7.
R1b1-P25 is found in Western Eurasia. Haplogroup
R1b1* is found in Africa at various frequencies. BerniellLee
et al (2009) found in their study that 5.2% carried
Rb1*. The frequency of R1b1* among the Bantu ranged
from 2-20. The bearers of R1b1* among the Pygmy
populations ranged from 1-25% (Berniell-Lee et al, 2009).
The frequency of R1b1 among Guinea-Bissau populations
was 12% (Carvalho et al,2010).
In relation to R-M269 in Africa I wrote:
quote:As a result, I did not attempt to decieve anyone about the frequency of R1 in Africa as the author implies.
Around 0.1 of Sub Saharan Africans carry R1b1b2. Wood
et al (2009) found that Khoisan (2.2%) and Niger-Congo
(0.4%) speakers carried the R-M269 y-chromosome.
The Khoisan also carry RM343 (R1b) and M 198 (R1a1)
(Naidoo et al., 2010) the archaeological and linguistic data
indicate the successful colonization of Asia by SubSaharan
Africans from Nubia 5-4kya (Winters, 2007,2008,
2010c). The archaeological evidence makes it clear that
around 4kya intercultural style artifacts connected Africa
and Eurasia (Winters, 2007,2010c).
In fact recent research on y-haplogroups in Africa suggest that R1-M269 is also widespread in Africa.
Above is a figure from Gonzalez et al. The Gonzalez et al article found that 10 out of 19 subjects in the study carried R1b1-P25 or M269. This is highly significant because it indicates that 53% of the R1 carriers in this study were M269, this finding is further proof of the widespread nature of this so-called Eurasian genes in Africa among populations that have not mated with Europeans.

quote:
Originally posted by the lioness,:
]However your information is obsolete due to recent analysis of Siberian DNA
Haplogroup R* was found in the remains of a Palaeolithic boy MA-1 (Mal'ta) near Lake Baikal in Siberia, dating to 24,000 years ago.
quote:Nobody said anything about Black or Caucasian. Stop being paranoid and making false allegations.
Originally posted by Clyde Winters:
It is bigotry because the Mal'ta boy was Black like the other Europeans in his day who lived at Pestera cu Oase; and you are trying to imply that he was Caucasian .
M207, like the rest of the R macrohaplogroup originated in Africa.
quote:Here's how you misquoted
Originally posted by Clyde Winters:
Above is a figure from Gonzalez et al. The Gonzalez et al article found that 10 out of 19 subjects in the study carried R1b1-P25 or M269. This is highly significant because it indicates that 53% of the R1 carriers in this study were M269,LOL. How can I be misquoting Gonzalez et al (2012), the figure is above and you can count the number of M269 subjects found in the study yourself.
.
quote:^^^ that is not the same as saying the people ofthe tiny country Equatorial Guinea (population 760,000 )
Originally posted by Clyde Winters:

quote:M269 is exremely sparse in Africa
The genetic landscape of Equatorial Guinea and the origin and migration routes of the Y chromosome haplogroup R-V88
Miguel González,
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3573200/#sup1
Characterisation of the male lineages of Equatorial Guinea
In the 112 samples analysed, we were able to identify 104 different haplotypes and 13 different haplogroups (Figure 1 and Supplementary Table S2). The majority of the Y-
SNP lineages found in this study (almost 80%) belong to haplogroup E, namely bearing the M2-derived allele, the most common haplogroup in sub-Saharan Africa Bantu populations. Apart from this haplogroup and five other chromosomes, all the remaining samples belong to haplogroup R, namely R1b1-P25, a lineage that is rare in Africa and is found mainly in Europe and Asia.
quote:--Cerný V1, Pereira L, Kujanová M, Vasíková A, Hájek M, Morris M, Mulligan CJ.
Abstract
The Soqotra archipelago is one of the most isolated landmasses in the world, situated at the mouth of the Gulf of Aden between the Horn of Africa and southern Arabia. The main island of Soqotra lies not far from the proposed southern migration route of anatomically modern humans out of Africa approximately 60,000 years ago (kya), suggesting the island may harbor traces of that first dispersal. Nothing is known about the timing and origin of the first Soqotri settlers. The oldest historical visitors to the island in the 15th century reported only the presence of an ancient population. We collected samples throughout the island and analyzed mitochondrial DNA and Y-chromosomal variation. We found little African influence among the indigenous people of the island. Although the island population likely experienced founder effects, links to the Arabian Peninsula or southwestern Asia can still be found. In comparison with datasets from neighboring regions, the Soqotri population shows evidence of long-term isolation and autochthonous evolution of several mitochondrial haplogroups. Specifically, we identified two high-frequency founder lineages that have not been detected in any other populations and classified them as a new R0a1a1 subclade. Recent expansion of the novel lineages is consistent with a Holocene settlement of the island approximately 6 kya.
quote:Really?
Originally posted by the lioness,:
quote:You must have a new definition of bigotry because Haplogroup R* was found in the remains of a Palaeolithic boy MA-1 (Mal'ta) near Lake Baikal in Siberia,
Originally posted by Troll Patrol # Ish Gebor:
quote:
Originally posted by the lioness,:
[qb]quote:However your information is obsolete due to recent analysis of Siberian DNA
Originally posted by Clyde Winters:
M207, like the rest of the R macrohaplogroup originated in Africa.
Haplogroup R* was found in the remains of a Palaeolithic boy MA-1 (Mal'ta) near Lake Baikal in Siberia, dating to 24,000 years ago.quote:Lol- this kind of bigotry is getting hilarious.
Originally posted by Quetzalcoatl:
Why woud such a fact be bigotry?
http://www.isogg.org/tree/ISOGG_HapgrpR.html
ISOGG
quote:
Y-DNA haplogroup R-M207 is believed to have arisen approximately 27,000 years ago in Asia. The two currently defined subclades are R1 and R2.
R1-M173 is estimated to have arisen during the height of the Last Glacial Maximum (LGM), about 18,500 years ago, most likely in southwestern Asia. The two most common descendant clades of R1 are R1a and R1b.
R1a-M420 is believed to have arisen on the Eurasian Steppe or the Indus Valley, and today is most frequently observed in eastern Europe and in western and central Asia. R1a1a1g-M458 is found at frequencies approaching or exceeding 30% in Eastern Europe.
R1b-M343 is believed to have arisen in southwest Asia and today its sublcades are bound in various distributions across Eurasia and Africa.
Paragroup R1b1* and R1b1c-V88 are found most frequently in SW Asia and Africa. The African examples are almost entirely within R1b1c and are associated with the spread of Chadic languages.
R1b1a-P297 is found throughout Eurasia. R1b1a1-M73 is observed most frequently in Asia, with low frequency of observation in Europe. R1b1a2-M269 is observed most frequently in Europe, especially western Europe, but with notable frequency in southwest Asia.
R1b1a2-M269 is estimated to have arisen approximately 4,000 to 8,000 years ago in southwest Asia and to have spread into Europe from there. The Atlantic Modal Haplotype, or AMH, is the most common STR haplotype in R1b1a2a1a-L11/PF6539/S127 and most European R1b1a2 belongs to R1b1a2a1a1-M405/S21/U106 or R1b1a2a1a2-P312/PF6547/S116.
R2-M479 is most often observed in Asia, especially on the Indian sub-continent and in central Asia.

quote:--Fulvio Cruciani et al.
‘‘Out of Africa’’ haplogroups.
All Y-clades that are not exclusively African belong to the macro-haplogroup CT, which is defined by mutations M168, M294 and P9.1 [14,31] and is subdivided into two major clades, DE and CF [1,14].
In a recent study [16], sequencing of two chromosomes belonging to haplogroups C and R, led to the identification of 25 new mutations, eleven of which were in the C-chromosome and seven in the R-chromosome.
Here, the seven mutations which were found to be shared by chromosomes of haplogroups C and R [16], were also found to be present in one DE sample (sample 33 in Table S1), and positioned at the root of macro-haplogroup CT (Figure 1 and Figure S1)
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3492319/figure/pone-0049170-g001/
Figure S1
Structure of the macro-haplogroup CT. For details on mutations see legend to Figure 1. Dashed lines indicate putative branchings (no positive control available). The position of V248 (haplogroup C2) and V87 (haplogroup C3) compared to mutations that define internal branches was not determined. Note that mutations V45, V69 and V88 have been previously mapped (Cruciani et al. 2010; Eur J Hum Genet 18∶800–807).
(TIF)
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3492319/bin/pone.0049170.s001.tif
quote:Use of Y Chromosome and Mitochondrial DNA Population Structure in Tracing Human Migrations
"haplogroup CF and DE molecular ancestors first evolved inside Africa and subsequently contributed as Y chromosome founders to pioneering migrations that successfully colonized Asia. While not proof, the DE and CF bifurcation (Figure 8d ) is consistent with independent colonization impulses possibly occurring in a short time interval."

quote:No we can not agree. The Khoisan were the Cro-Magnon people. They came from Africa and probably carried this haplogroup to Europe. They also may have introduced the haplogroup to North America since it was the Khoisan who took the Solutrean culture to North America.
Originally posted by the lioness,:
quote:
Originally posted by the lioness,:
]However your information is obsolete due to recent analysis of Siberian DNA
Haplogroup R* was found in the remains of a Palaeolithic boy MA-1 (Mal'ta) near Lake Baikal in Siberia, dating to 24,000 years ago.quote:Nobody said anything about Black or Caucasian. Stop being paranoid and making false allegations.
Originally posted by Clyde Winters:
It is bigotry because the Mal'ta boy was Black like the other Europeans in his day who lived at Pestera cu Oase; and you are trying to imply that he was Caucasian .
M207, like the rest of the R macrohaplogroup originated in Africa.
I just put up the ISOGG stating that R originates in Asia far prior to V88 or M269
Siberia is a lot farther away from Africa than Europe is from Africa
and Siberia is on the same landamss as Europe while Africa isn't.
So now we can agree that R originates in North East Asia 24kya rather than Africa and you were wrong. Can we go home now?
quote:I did not say 53% of the entire population of the country was M269, and you know that.I was only talking about the people who carry haplogroup R1.
Originally posted by the lioness,:
quote:Here's how you misquoted
Originally posted by Clyde Winters:
Above is a figure from Gonzalez et al. The Gonzalez et al article found that 10 out of 19 subjects in the study carried R1b1-P25 or M269. This is highly significant because it indicates that 53% of the R1 carriers in this study were M269,LOL. How can I be misquoting Gonzalez et al (2012), the figure is above and you can count the number of M269 subjects found in the study yourself.
.
above you are saying
"10 out of 19 subjects in the study carried R1b1-P25 or M269. This is highly significant because it indicates that 53% of the R1 carriers in this study were M269"
Then you turned this into a frequency chart, your own below
quote:^^^ that is not the same as saying the people ofthe tiny country Equatorial Guinea (population 760,000 )
Originally posted by Clyde Winters:

have frequency averages of 53%
That is not frequency - that is the proportion who were M269 carriers
-but does not indicate how much M269 was present for each individuals Y DNA profile
That is why they don't list or talk about percentage on those figures
quote:M269 is exremely sparse in Africa
The genetic landscape of Equatorial Guinea and the origin and migration routes of the Y chromosome haplogroup R-V88
Miguel González,
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3573200/#sup1
Characterisation of the male lineages of Equatorial Guinea
In the 112 samples analysed, we were able to identify 104 different haplotypes and 13 different haplogroups (Figure 1 and Supplementary Table S2). The majority of the Y-
SNP lineages found in this study (almost 80%) belong to haplogroup E, namely bearing the M2-derived allele, the most common haplogroup in sub-Saharan Africa Bantu populations. Apart from this haplogroup and five other chromosomes, all the remaining samples belong to haplogroup R, namely R1b1-P25, a lineage that is rare in Africa and is found mainly in Europe and Asia.
CLYDE THEY IDENTIFIED 13 DIFFERENT HAPLOGROUPS M269 IS JUST ONE OF THEM and they did not compile the different frequencies of each haplogroup for each individual nor as a whole, merely that they carried SOME percentage of these 13 Haplogroups

quote:this chart made by Clyde is incorrect
Originally posted by Clyde Winters:

quote:
Genetic and Demographic Implications of the Bantu Expansion: Insights from Human Paternal Lineages
Biernell-Lee et al 2009
On the whole, most of the samples belonged to previously described African lineages especially common in sub-Saharan Africa. Specifically, most of these lineages have been associated either with Bantu-speaking people—E1b1a (E3a according to The Y Chromosome Consortium 2002), B2a, and E2—or to Pygmy populations (haplogroup B2b). We also observed traces of haplogroups A, E*, E1a, and E1b1b1a (E3b1 according to The Y Chromosome Consortium 2002), which are found at low frequencies across the African continent (Underhill et al. 2000, 2001; Cruciani et al. 2002; Wood et al. 2005). Interestingly, almost 5% of the individuals here analyzed belonged to Eurasian haplogroup R1b1*.
Haplogroup E1b1a, previously proposed as being a marker of the Bantu expansion (Passarino et al. 1998; Scozzari et al. 1999; Underhill et al. 2001; Wood et al. 2005), was the most frequent haplogroup in our sample set (76%), reaching a frequency of almost 80% in agriculturalists and 28% in Pygmy samples (fig. 1).
http://mbe.oxfordjournals.org/content/26/7/1581/F1.large.jpg


quote:
Originally posted by Troll Patrol # Ish Gebor:
quote:LOL at Low Level Thinker.
Originally posted by the lioness,:
Quiet jackass I'm not talking to you, xyyman is talking about Hgs not alleles.
His concept is that white Europeans are depigmenated Africans, not Central Asians and their ancestors lived in Africa less than 5,000 years ago so that's the "we" you are a part of.
To say that that chart is stupid means you are stupid
Hg's are based on alleles. Speaking of quite a jackass.![[Big Grin]](biggrin.gif)
quote:LOL. That's what researcher do, you take primary data and make new knowledge based on it.
Originally posted by the lioness,:
quote:this chart made by Clyde is incorrect
Originally posted by Clyde Winters:

example, Biernell-Lee's 2009 paper >>
quote:
Genetic and Demographic Implications of the Bantu Expansion: Insights from Human Paternal Lineages
Biernell-Lee et al 2009
On the whole, most of the samples belonged to previously described African lineages especially common in sub-Saharan Africa. Specifically, most of these lineages have been associated either with Bantu-speaking people—E1b1a (E3a according to The Y Chromosome Consortium 2002), B2a, and E2—or to Pygmy populations (haplogroup B2b). We also observed traces of haplogroups A, E*, E1a, and E1b1b1a (E3b1 according to The Y Chromosome Consortium 2002), which are found at low frequencies across the African continent (Underhill et al. 2000, 2001; Cruciani et al. 2002; Wood et al. 2005). Interestingly, almost 5% of the individuals here analyzed belonged to Eurasian haplogroup R1b1*.
Haplogroup E1b1a, previously proposed as being a marker of the Bantu expansion (Passarino et al. 1998; Scozzari et al. 1999; Underhill et al. 2001; Wood et al. 2005), was the most frequent haplogroup in our sample set (76%), reaching a frequency of almost 80% in agriculturalists and 28% in Pygmy samples (fig. 1).
http://mbe.oxfordjournals.org/content/26/7/1581/F1.large.jpg
As we see here Biernell-Lee reports 0% M269 in the right column of the chart. Insiead R1b1* aka R-P25
And note the numbers are not frequencies. They are just the number of individuals in each group that carried the haplogroup
For instance look again at the quote on E1b1>
E1b1a, previously proposed as being a marker of the Bantu expansion , was the most frequent haplogroup in our sample set (76%), reaching a frequency of almost 80% in agriculturalists and 28% in Pygmy samples
^^^ 76% of the individuals carried E1b1a
But how much on average did those that carried E1b1a ?
Was it 2% or 92%
In this case 80%
That is the frequency, a separate issue as stated
Now let's look at Cruciani 2010 as regards M269 in Africa
Here the figures ARE frequency >
on the far right, haplogroup M269
highest frequency perceatage 2.4% Semitic Egyptians from Baharia
by contrast at the bottom of the chart Western Europeans 57.8%
Perhaps Clyde has R-V69 confused with R-M269 (?)
Clyde Winters is misinterpreting information and making his own charts based on it
quote:Yes, and they are NOT 53% of the haplogroups. If you look at Table 1 in the paper, p. 326, Gonzales gives you both the raw numbers and the proper percentages:
Originally posted by Clyde Winters:
quote:LOL. How can I be misquoting Gonzalez et al (2012), the figure is above and you can count the number of M269 subjects found in the study yourself.
Originally posted by Quetzalcoatl:
quote:In a later post I show that you are misinterpreting/misquoting these sources. R-M269 is NOT African but European and the only place with significant R1 is Cameroon where the haplotype is R-V88.
Originally posted by Clyde Winters:
Klyosov in criticizing my article exagerates what I wrote. He implies that I was trying to decieve the readers about the frequency of R1 in Africa. This is false.
I specifically stated the frequency of R1 among African populations throughout my 2011 paper.
quote:As you can see I did admit that only 5.2% of the pygmy carried R1.
Y-chromosome R1 is found throghout Africa. The pristine
form of R1-M173 is only found in Africa (Coia et al,
2005; Cruciani et al, 2002, 2010). The age of ychromosome
R is 27ky. Most researchers believe that
R(M173) is 18.5 ky.There is a great diversity of the
macrohaplogroup R in Africa (See Figure 1). Ychromosome
R is characterized by M207/V45. The V45
mutation is found among African populations ( Cruciani et
al ,2010). ISOGG 2010 Y-DNA haplogroup tree makes it
clear that V45 is phylogenetically equivalent to M207.The
most common R haplogroup in Africa is R1 (M173). The
predominant haplogroup is R1b (Berniell-Lee et al,2009;
Coia et al, 2005; Winters, 2010b; Wood et al, 2009).
Cruciani et al (2010) discovered new R1b mutations
including V7, V8, V45, V69, and V88. Geography appears
to play a significant role in the distribution of haplogroup
R in Africa. Cruciani et al (2010) has renamed the R*-
M173 (R P-25) in Africa V88. The TMRCA of V88 was
9200-5600 kya (Cruciani et al, 2010).
Y-chromosome V88 (R1b1a) has its highest frequency
among Chadic speakers, while the carriers of V88 among
Niger-Congo speakers (predominately Bantu people)
range between 2-66% ( Cruciani et al, 2010; Bernielle-Lee
et al, 2009). Haplogroup V88 includes the mutations M18,
V35 and V7. Cruciani et al (2010) revealed that R-V88 is
also carried by Eurasians including the distinctive
mutations M18, V35 and V7.
R1b1-P25 is found in Western Eurasia. Haplogroup
R1b1* is found in Africa at various frequencies. BerniellLee
et al (2009) found in their study that 5.2% carried
Rb1*. The frequency of R1b1* among the Bantu ranged
from 2-20. The bearers of R1b1* among the Pygmy
populations ranged from 1-25% (Berniell-Lee et al, 2009).
The frequency of R1b1 among Guinea-Bissau populations
was 12% (Carvalho et al,2010).
In relation to R-M269 in Africa I wrote:
quote:As a result, I did not attempt to decieve anyone about the frequency of R1 in Africa as the author implies.
Around 0.1 of Sub Saharan Africans carry R1b1b2. Wood
et al (2009) found that Khoisan (2.2%) and Niger-Congo
(0.4%) speakers carried the R-M269 y-chromosome.
The Khoisan also carry RM343 (R1b) and M 198 (R1a1)
(Naidoo et al., 2010) the archaeological and linguistic data
indicate the successful colonization of Asia by SubSaharan
Africans from Nubia 5-4kya (Winters, 2007,2008,
2010c). The archaeological evidence makes it clear that
around 4kya intercultural style artifacts connected Africa
and Eurasia (Winters, 2007,2010c).
In fact recent research on y-haplogroups in Africa suggest that R1-M269 is also widespread in Africa.
Above is a figure from Gonzalez et al. The Gonzalez et al article found that 10 out of 19 subjects in the study carried R1b1-P25 or M269. This is highly significant because it indicates that 53% of the R1 carriers in this study were M269, this finding is further proof of the widespread nature of this so-called Eurasian genes in Africa among populations that have not mated with Europeans.
.

.
quote:LOL. You're really dumb. 19 R1 subjects in study. 10 of the subjects carry M269. 10/19= 53%.
Originally posted by Quetzalcoatl:
quote:Yes, and they are NOT 53% of the haplogroups. If you look at Table 1 in the paper, p. 326, Gonzales gives you both the raw numbers and the proper percentages:
Originally posted by Clyde Winters:
quote:LOL. How can I be misquoting Gonzalez et al (2012), the figure is above and you can count the number of M269 subjects found in the study yourself.
Originally posted by Quetzalcoatl:
quote:In a later post I show that you are misinterpreting/misquoting these sources. R-M269 is NOT African but European and the only place with significant R1 is Cameroon where the haplotype is R-V88.
Originally posted by Clyde Winters:
Klyosov in criticizing my article exagerates what I wrote. He implies that I was trying to decieve the readers about the frequency of R1 in Africa. This is false.
I specifically stated the frequency of R1 among African populations throughout my 2011 paper.
quote:As you can see I did admit that only 5.2% of the pygmy carried R1.
Y-chromosome R1 is found throghout Africa. The pristine
form of R1-M173 is only found in Africa (Coia et al,
2005; Cruciani et al, 2002, 2010). The age of ychromosome
R is 27ky. Most researchers believe that
R(M173) is 18.5 ky.There is a great diversity of the
macrohaplogroup R in Africa (See Figure 1). Ychromosome
R is characterized by M207/V45. The V45
mutation is found among African populations ( Cruciani et
al ,2010). ISOGG 2010 Y-DNA haplogroup tree makes it
clear that V45 is phylogenetically equivalent to M207.The
most common R haplogroup in Africa is R1 (M173). The
predominant haplogroup is R1b (Berniell-Lee et al,2009;
Coia et al, 2005; Winters, 2010b; Wood et al, 2009).
Cruciani et al (2010) discovered new R1b mutations
including V7, V8, V45, V69, and V88. Geography appears
to play a significant role in the distribution of haplogroup
R in Africa. Cruciani et al (2010) has renamed the R*-
M173 (R P-25) in Africa V88. The TMRCA of V88 was
9200-5600 kya (Cruciani et al, 2010).
Y-chromosome V88 (R1b1a) has its highest frequency
among Chadic speakers, while the carriers of V88 among
Niger-Congo speakers (predominately Bantu people)
range between 2-66% ( Cruciani et al, 2010; Bernielle-Lee
et al, 2009). Haplogroup V88 includes the mutations M18,
V35 and V7. Cruciani et al (2010) revealed that R-V88 is
also carried by Eurasians including the distinctive
mutations M18, V35 and V7.
R1b1-P25 is found in Western Eurasia. Haplogroup
R1b1* is found in Africa at various frequencies. BerniellLee
et al (2009) found in their study that 5.2% carried
Rb1*. The frequency of R1b1* among the Bantu ranged
from 2-20. The bearers of R1b1* among the Pygmy
populations ranged from 1-25% (Berniell-Lee et al, 2009).
The frequency of R1b1 among Guinea-Bissau populations
was 12% (Carvalho et al,2010).
In relation to R-M269 in Africa I wrote:
quote:As a result, I did not attempt to decieve anyone about the frequency of R1 in Africa as the author implies.
Around 0.1 of Sub Saharan Africans carry R1b1b2. Wood
et al (2009) found that Khoisan (2.2%) and Niger-Congo
(0.4%) speakers carried the R-M269 y-chromosome.
The Khoisan also carry RM343 (R1b) and M 198 (R1a1)
(Naidoo et al., 2010) the archaeological and linguistic data
indicate the successful colonization of Asia by SubSaharan
Africans from Nubia 5-4kya (Winters, 2007,2008,
2010c). The archaeological evidence makes it clear that
around 4kya intercultural style artifacts connected Africa
and Eurasia (Winters, 2007,2010c).
In fact recent research on y-haplogroups in Africa suggest that R1-M269 is also widespread in Africa.
Above is a figure from Gonzalez et al. The Gonzalez et al article found that 10 out of 19 subjects in the study carried R1b1-P25 or M269. This is highly significant because it indicates that 53% of the R1 carriers in this study were M269, this finding is further proof of the widespread nature of this so-called Eurasian genes in Africa among populations that have not mated with Europeans.
.

.
number percentage
V88 Ribia* 9 (8.04%)
M269 R1b1b2 10 (8.93)
You are still misquoting the true values given in your sources.
quote:Yes, and they are NOT 53% of the haplogroups. If you look at Table 1 in the paper, p. 326, Gonzales gives you both the raw numbers and the proper percentages:
Originally posted by Clyde Winters:
[

.
quote:There is no conspiracy. Take a look at Phylotree http://www.phylotree.org/Y/tree/index.htm practically every line has changed names as new haplotypes are discovered. You claim to be a scientist and lecture others on what science is, yet you make the statement : "They can change whatever they wish. I still believe it originated in Africa,". This is religion not science refusing to look at evidence.
Originally posted by Clyde Winters:
quote:LOL. They can change whatever they wish. I still believe it originated in Africa.
Originally posted by the lioness,:
quote:that is incorrect
Originally posted by Clyde Winters:
R-M207 is the same as V45.
ISOGG made an error or correction on this
http://www.isogg.org/tree/ISOGG_HapgrpR10.html
quote:^^^ here they show V5 as an allele of 207
R M207/UTY2, P224, P227, P229, P232, P280,
P285, S4, S8, S9, V45
However on the same page:
quote:^^^ added SNPs > reference Cruciani et al (2010) >>>
Corrections/Additions made since 1 January 2010:
Added the following SNPs L165, L193, L196, L226, P89.2, V7, V8, V35, V45, V69, V88 and Cruciani et al (2010) on 12 January 2010
quote:^^^ this is the sole and primary source about V45

We resequenced about 0.15 Mb of the MSY for each of the four R1b subjects and found six new mutations (V7, V8, V35, V45, V69, and V88). The V45 mutation is phylogenetically equivalent to M173. Among the other five mutations, V88 defines a new monophyletic clade (R-V88 or R1b1a), which includes haplogroups R-M18 (R1b1a1, formerly R1b1a), R-V8 (R1b1a2), R-V35 (R1b1a3, further subdivided by the V7 mutation to R1b1a3* and R1b1a3a), and R-V69 (R1b1a4) (Figure 1).
-- Human Y chromosome haplogroup R-V88: a paternal genetic record of early mid Holocene trans-Saharan connections and the spread of Chadic languages
Fulvio Cruciani, 2010
The chart above is for R1
However R-M207 is ancestor to R1
That's the paragroup, the R* root before R1 came into existence
quote:LOL. You're really dumb. 19 R1 subjects in study. 10 of the subjects carry M269. 10/19= 53%.
Originally posted by Quetzalcoatl:
quote:Yes, and they are NOT 53% of the haplogroups. If you look at Table 1 in the paper, p. 326, Gonzales gives you both the raw numbers and the proper percentages:
Originally posted by Clyde Winters:
[

.
number percentage
V88 Ribia* 9 (8.04%)
M269 R1b1b2 10 (8.93)
You are still misquoting the true values given in your sources.

quote:^^^ What he's saying is correct, the only correction is that it's Figure 1 not Table 1.
Originally posted by Quetzalcoatl:
quote:Yes, and they are NOT 53% of the haplogroups. If you look at Table 1 in the paper, p. 326, Gonzales gives you both the raw numbers and the proper percentages:
Originally posted by Clyde Winters:

number percentage
V88 Ribia* 9 (8.04%)
M269 R1b1b2 10 (8.93)
You are still misquoting the true values given in your sources.

quote:Clyde Hassan 2008 reported that Sudanese Fulani carry R-M173 (x P25) at a 54% frequency
The phylogeographic profile of R1*-M173 supports this ancient migration of Kushites from Africa to Eurasia as suggested by the Classical writers. This expansion of Kushites into Eurasia probably took place over 4 kya.
--Clyde Winters
The Kushite Spread of Haplogroup R1*-M173 from Africa to Eurasia
quote:You base your argument on contemporary frequencies of R1a and R1b among Europeans. As has been shown repeatedly on this forum white Europeans did not originate in Europe, they come from Central Asia as a result the frequency of the R haplogroup among this population does not reflect the age of haplogroup R.
Originally posted by the lioness,:
quote:Clyde Hassan 2008 reported that Sudanese Fulani carry R-M173 (x P25) at a 54% frequency
The phylogeographic profile of R1*-M173 supports this ancient migration of Kushites from Africa to Eurasia as suggested by the Classical writers. This expansion of Kushites into Eurasia probably took place over 4 kya.
--Clyde Winters
The Kushite Spread of Haplogroup R1*-M173 from Africa to Eurasia
You say an expansion of Kushites into Eurasia probably took place over 4 kya.
But R-M173 is estimated to be 12,500–25,700 years old.
Assuming that Sudanese Fulani are descendants of the Kushites are you saying that there was no R1a outside of Africa until around 4,000 years ago?
That doesn't make sense
In Europe, the highest frequencies are found in Central and Eastern Europe. Today it is found at its highest levels in Tajiks (64%), Kyrgyz (63%), Poland and Hungary (56%–60%), Ukraine (44-54%) High haplotype diversity was detected in Poland
Among the caste groups of India high percentages of this haplogroup are found in West Bengal Brahmins(72%) to the east, Konkanastha Brahmins(48%) to the west, Khatris(67%) in north and Iyenger(31%) Brahmins of south. It has also been found in several South Indian Dravidian-speaking Adivasis including the Chenchu(26%), the Valmikis of Andhra Pradesh and the Kallar of Tamil Nadu
Absolute dating methods suggest that this marker is 10–15,000 years old, and the microsatellite diversity is greatest in southern Asia.
The modern population of South Asia has the highest level of diversity of the gene making it the likeliest location of its origin
My guess is that the Indians carried this Haplogroup to both Europe and Africa
The ancestor of this Haplogroup is R* found in 24 kya Siberia remains
R-M173 is at minimum over 12,500 years old. It has a wide range in Eurasia so it could not have first arrived a mere 4000 or so years ago




 Given the lack of life in the caves these first caucasians probably ate each other and suffered many bad things.
Given the lack of life in the caves these first caucasians probably ate each other and suffered many bad things.


quote:
Originally posted by Mike111:
Twenty nine ivory female figures were found on the open site of Mal'ta. They differ from contemporary representations in Russia, Central and Western Europe in that they are shown clothed rather than nude, most have faces and the body shapes are straight or tapering below large, round heads. Some are also perforated to be worn as pendants.
.
However many are Steatopygic indicating Khoisan/Grimaldi ethnicity.



quote:[/QB][/QUOTE]
Originally posted by Narmerthoth:

Stone Age Tunnels are man created a massive network of underground tunnels crisecrossing Europe from Scotland to Turkey , a new book on the Ancient superhighways has claimed. German archaeologist, said evidence of the tunnels has been found hundreds of Neolithic settlements all over the continent. Seemingly never ending series of underground tunnels are confirmed to be man-made , and the architecture boggles even the most sophisticated designers.
German archaeologist Dr Heinrich Kusch said evidence of the tunnels has been found under hundreds of Neolithic settlements all over the continent. In his book - Secrets Of The Underground Door To An Ancient World (German title: Tore zur Unterwelt) - he says that the fact so many have survived after 12,000 years shows that the original tunnel network must have been enormous.
In his book, he notes that chapels were often built by the entrances perhaps because the Church were afraid of the heathen legacy the tunnels might have represented, and wanted to negate their influence.

Not for the claustrophobic: Most of the tunnels are just 70cm wide - just wide enough for a person to slowly wriggle through
quote:Since the Proto-Europeans in western Eurasia, like those living at Grotte de Marche remaind in their self imposed imprisonment underground, they did not interact with the Black Europeans. As a result, they cannot be considered Europeans because they were either unable, or afraid to exit the caves of western Eurasia.
Santorini's volcanic activity during the past 2-500,000 years has been dominated by very large explosive eruptions at intervals of few tens of thousands of years. The most recent of this type occurred at around 1613 BC and is known as the so-called Minoan eruption. The late Bronze Age eruption, one of the biggest known volcanic explosions in younger time,- and one of the most studied, but still most mysterious eruptions of all time-, devastated not only Santorini, but had a deep impact on the whole of the Eastern Mediterranean. Perhaps it even had serious world-wide effects and changed history.
Still today, one can see its deposits, the characteristic, tens of meters thick layer of white pumice and ash that blankets most of the surface of the island group. The eruption changed the shape of the island itself dramatically: it is now believed that before the eruption, it had the shape of an almost complete ring that enclosed an earlier, shallower caldera. Then, large sections of island collapsed into the emptied magma chamber after the eruption, literally disappearing under the sea. The ring-island was breached to the W and NW, and the caldera was significantly widened and deepened.
The Minoan eruption devastated the rich, highly developed economic center, that Santorini was at that time. Since 1969, intense archaeological excavations have brought to light an important Cycladic/Minoan town which had been buried beneath the volcanic ash for almost 4000 years. Although it appears that people had time to evacuate their island in time before the eruption, carrying most of their goods with them, the findings from Akrotiri are impressive: especially, they include well-preserved and magnificent wall paintings, ceramics and other objects. Thanks to the work of numerous archaeologists, a new light was thrown on an important prehistoric period and culture. The spectacular discovery even induced continuing speculations that relate the volcanic destruction of Santorini to the legend of the sunken Atlantis.
See:
http://www.volcanodiscovery.com/santorini.html
quote:That is because the origin was in Africa. There is where the basal is at.
Originally posted by xyyman:
To those who are slow…er. And needs things to be simplified. Busby is stating that R1b-269 did NOT come from the Middle East or Steppes. Yes, the Yamanya carried R1b but as the same authors clearly stated, the R1b of the Yamanya was of a different “type”. Which corroborates what Busby disclosed. Western European R1b-M269 did NOT come from the Asian Steppes.
Busby provided data as such. Unfortunately he did NOT provide an origin. He just discredited the notion of an “Eastern” source.
quote:--Hong Shi et al. 2008:
The Y chromosome Alu polymorphism (YAP, also called M1) defines the deep-rooted haplogroup D/E of the global Y-chromosome phylogeny [1]. This D/E haplogroup is further branched into three sub-haplogroups DE*, D and E (Figure 1). The distribution of the D/E haplogroup is highly regional, and the three subgroups are geographically restricted to certain areas, therefore informative in tracing human prehistory (Table 1). The sub-haplogroup DE*, presumably the most ancient lineage of the D/E haplogroup was only found in Africans from Nigeria [2], supporting the "Out of Africa" hypothesis about modern human origin. The sub-haplogroup E (E-M40), defined by M40/SRY4064 and M96, was also suggested originated in Africa [3-6], and later dispersed to Middle East and Europe about 20,000 years ago [3,4]. Interestingly, the sub-haplogroup D defined by M174 (D-M174) is East Asian specific with abundant appearance in Tibetan and Japanese (30–40%), but rare in most of other East Asian populations and populations from regions bordering East Asia (Central Asia, North Asia and Middle East) (usually less than 5%) [5-7]. Under D-M174, Japanese belongs to a separate sub-lineage defined by several mutations (e.g. M55, M57 and M64 etc.), which is different from those in Tibetans implicating relatively deep divergence between them [1]. The fragmented distribution of D-M174 in East Asia seems not consistent with the pattern of other East Asian specific lineages, i.e. O3-M122, O1-M119 and O2-M95 under haplogroup O [8,9].
quote:Haplogroup DE* in Guinea-Bissau:
Further refinement awaits the finding of new markers especially within paragroup E3a*-M2. The microsatellite profile of the DE* individual is one mutational step away from the allelic state described for Nigerians (DYS390*21, DYS388 not tested; [37], therefore suggesting a common ancestry but not elucidating the phylogenetics.
quote:Haplogroup DE* in Nigerians:
There has been considerable debate on the geographic origin of the human Y chromosome Alu polymor- phism (YAP). Here we report a new, very rare deep-rooting haplogroup within the YAP clade, together with data on other deep-rooting YAP clades. The new haplogroup, found so far in only five Nigerians, is the least-derived YAP haplogroup according to currently known binary markers. However, because the interior branching order of the Y chromosome genealogical tree remains unknown, it is impossible to impute the origin of the YAP clade with certainty. We discuss the problems presented by rare deep-rooting lineages for Y chromosome phylogeography.
quote:--Fulvio Cruciani et al.
‘‘Out of Africa’’ haplogroups.
All Y-clades that are not exclusively African belong to the macro-haplogroup CT, which is defined by mutations M168, M294 and P9.1 [14,31] and is subdivided into two major clades, DE and CF [1,14].
In a recent study [16], sequencing of two chromosomes belonging to haplogroups C and R, led to the identification of 25 new mutations, eleven of which were in the C-chromosome and seven in the R-chromosome.
Here, the seven mutations which were found to be shared by chromosomes of haplogroups C and R [16], were also found to be present in one DE sample (sample 33 in Table S1), and positioned at the root of macro-haplogroup CT (Figure 1 and Figure S1)
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3492319/figure/pone-0049170-g001/
Figure S1
Structure of the macro-haplogroup CT. For details on mutations see legend to Figure 1. Dashed lines indicate putative branchings (no positive control available). The position of V248 (haplogroup C2) and V87 (haplogroup C3) compared to mutations that define internal branches was not determined. Note that mutations V45, V69 and V88 have been previously mapped (Cruciani et al. 2010; Eur J Hum Genet 18∶800–807).
(TIF)
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3492319/bin/pone.0049170.s001.tif
quote:Use of Y Chromosome and Mitochondrial DNA Population Structure in Tracing Human Migrations
"haplogroup CF and DE molecular ancestors first evolved inside Africa and subsequently contributed as Y chromosome founders to pioneering migrations that successfully colonized Asia. While not proof, the DE and CF bifurcation (Figure 8d ) is consistent with independent colonization impulses possibly occurring in a short time interval."
quote:The more I find on this the more I side with you on this.
Originally posted by Clyde Winters:
quote:You base your argument on contemporary frequencies of R1a and R1b among Europeans. As has been shown repeatedly on this forum white Europeans did not originate in Europe, they come from Central Asia as a result the frequency of the R haplogroup among this population does not reflect the age of haplogroup R.
Originally posted by the lioness,:
quote:Clyde Hassan 2008 reported that Sudanese Fulani carry R-M173 (x P25) at a 54% frequency
The phylogeographic profile of R1*-M173 supports this ancient migration of Kushites from Africa to Eurasia as suggested by the Classical writers. This expansion of Kushites into Eurasia probably took place over 4 kya.
--Clyde Winters
The Kushite Spread of Haplogroup R1*-M173 from Africa to Eurasia
You say an expansion of Kushites into Eurasia probably took place over 4 kya.
But R-M173 is estimated to be 12,500–25,700 years old.
Assuming that Sudanese Fulani are descendants of the Kushites are you saying that there was no R1a outside of Africa until around 4,000 years ago?
That doesn't make sense
In Europe, the highest frequencies are found in Central and Eastern Europe. Today it is found at its highest levels in Tajiks (64%), Kyrgyz (63%), Poland and Hungary (56%–60%), Ukraine (44-54%) High haplotype diversity was detected in Poland
Among the caste groups of India high percentages of this haplogroup are found in West Bengal Brahmins(72%) to the east, Konkanastha Brahmins(48%) to the west, Khatris(67%) in north and Iyenger(31%) Brahmins of south. It has also been found in several South Indian Dravidian-speaking Adivasis including the Chenchu(26%), the Valmikis of Andhra Pradesh and the Kallar of Tamil Nadu
Absolute dating methods suggest that this marker is 10–15,000 years old, and the microsatellite diversity is greatest in southern Asia.
The modern population of South Asia has the highest level of diversity of the gene making it the likeliest location of its origin
My guess is that the Indians carried this Haplogroup to both Europe and Africa
The ancestor of this Haplogroup is R* found in 24 kya Siberia remains
R-M173 is at minimum over 12,500 years old. It has a wide range in Eurasia so it could not have first arrived a mere 4000 or so years ago
Granted, Khoisan probably introduced haplogroup R into Europe as early as 24kya. But Khoisan were later replaced by the Anu (Pygmies) and finally the Kushites. This means that the haplogroups carried by these later populations would have influenced the genetic landscape of Eurasia.
Since caucasians and mongoloid people do not appear in the archeological record until 2000 BC, and we see the replacement of Black populations in Europe and Central Asia after 1500 by whites (as evidenced by the People of the Sea). By this time the Great Flood had destroyed most Anu centers in Eurasia, and the Kushites had resettled many centers formerly occupied by the Anu.
As a result, Europeans carrying the R haplogroups today are the result of mating with the Kushites, who they met on their way from Central Asia into Europe. The only other way we can account for the R haplogroup among whites, is that when they exited their cave homeland in the caucasus mountains the Proto-white people were carrying the R haplogroup of their Khoisan ancestors. If this view is correct, the R haplogroup was the main haplogroup carried by whites during their sojurn in the caves .
quote:--Pavel M. DOLUKHANOV
The population of AMH spreading in the eastern direction included “softened” Mongoloid elements. The “dialectal continuum” consisting of Proto-Uralic, Proto-Altaic and Palaeo-Siberian- related languages formed the principal communication media of Early Modern Humans in northern Eurasia.
quote:correct, according to what is currently known Haplogroup R did not originate in Africa
Originally posted by Clyde Winters:
The earliest evidence of the R haplogroup comes from Mal’ta Siberia.
quote:Apart from the misleading 53% on Equatorial Guinea there are other problems with the figures you cite. The entire table is titled “African Carriers of R-M269” but in reality R-M269 is extremely rare in Africa. Instead what you are listing are amounts of R1b* (also called R-P25*, R-M343, R-V88). A part of the problem is the unsettled state of the nomenclature and the fact that over the last 10 years different papers have used different names.
Originally posted by Clyde Winters:
.

.
quote:But in Appendix A What we see is a high frequency of R-P25 (R1b1*) in Cameroon But there is ZERO R-M269.
While African haplogroup R chromosomes are generally quite rare, R-P25* chromosomes are found at remarkably high frequencies in northern Cameroon (60.7–94.7%).
quote:LOL. You admit that the figures in the Table are recorded accurately. And then cite other figures that have nothing to do with M269.
Originally posted by Quetzalcoatl:
quote:Apart from the misleading 53% on Equatorial Guinea there are other problems with the figures you cite. The entire table is titled “African Carriers of R-M269” but in reality R-M269 is extremely rare in Africa. Instead what you are listing are amounts of R1b* (also called R-P25*, R-M343, R-V88). A part of the problem is the unsettled state of the nomenclature and the fact that over the last 10 years different papers have used different names.
Originally posted by Clyde Winters:
.

.
For example in Berniell-Lee:
Your table says R-M269 2-20% in Bantu and in Pygmies 5%, but on
p. 1583 what is listed as present is haplotype R-P25 (R1b1*) with 14% frequency in the Bantu. However, there is zero haplotype R-M269 in either Bantu, or Pygmies
In Wood:
You say R-M269 is present in 2.2 % of the Khoisan. On p. 872 she saysquote:But in Appendix A What we see is a high frequency of R-P25 (R1b1*) in Cameroon But there is ZERO R-M269.
While African haplogroup R chromosomes are generally quite rare, R-P25* chromosomes are found at remarkably high frequencies in northern Cameroon (60.7–94.7%).
Your cite of Hirbo at 6% R-M269 in the Khoisan is correct.
p. 199 Appendix 6a (ii) part III: Frequency of Y chromosome lineages in Africa, Mediterranean and the Near East
what is listed at a very low level are R1b*[R-V88]and R1b3 (Really R-M269)
The frequencies above 10% of R1b3 (R-M269) listed are:
Algerians (10.8), Iraquis(10.80),Turks(14.7), Greeks(17.4), Albanians(18.5), Sicilians(24.7), Sardinians(19.2), Spanish(65.6), Portuguese(58.4), Sao Tome(10.1).
AND Tsumkwe Namibian Ju’hoanse San= 6%
Berniell-Lee, G. et al. “Genetic and Demographic Implications of the Bantu Expansion: Insights from Human Paternal Lineages,” Mol. Biol. Evol. 26(7): 1581–1589.
Wood, E. T., et al. 2005 “Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes,” European Journal of Human Genetics 13: 867–876
Hirbo, J.H. 2011 Complex Genetic History of East African Human Populations PhD Dissertation, Department of Biology, University of Maryland
quote:Can you read? The table purports to show the presence of R-M269 in Africa. I just quoted your sources that there is NO, NONE, ZERO R-M269 in Africa (except a little in Khoisan, and North Africa). You, as usual, did not quote your sources accurately. What I show is that the sources were talking about a different haplotype R-V88 (also named R1b1*) and that even there your numbers were not accurate.
Originally posted by Clyde Winters:
quote:LOL. You admit that the figures in the Table are recorded accurately. And then cite other figures that have nothing to do with M269.
Originally posted by Quetzalcoatl:
quote:Apart from the misleading 53% on Equatorial Guinea there are other problems with the figures you cite. The entire table is titled “African Carriers of R-M269” but in reality R-M269 is extremely rare in Africa. Instead what you are listing are amounts of R1b* (also called R-P25*, R-M343, R-V88). A part of the problem is the unsettled state of the nomenclature and the fact that over the last 10 years different papers have used different names.
Originally posted by Clyde Winters:
.

.
For example in Berniell-Lee:
Your table says R-M269 2-20% in Bantu and in Pygmies 5%, but on
p. 1583 what is listed as present is haplotype R-P25 (R1b1*) with 14% frequency in the Bantu. However, there is zero haplotype R-M269 in either Bantu, or Pygmies
In Wood:
You say R-M269 is present in 2.2 % of the Khoisan. On p. 872 she saysquote:But in Appendix A What we see is a high frequency of R-P25 (R1b1*) in Cameroon But there is ZERO R-M269.
While African haplogroup R chromosomes are generally quite rare, R-P25* chromosomes are found at remarkably high frequencies in northern Cameroon (60.7–94.7%).
Your cite of Hirbo at 6% R-M269 in the Khoisan is correct.
p. 199 Appendix 6a (ii) part III: Frequency of Y chromosome lineages in Africa, Mediterranean and the Near East
what is listed at a very low level are R1b*[R-V88]and R1b3 (Really R-M269)
The frequencies above 10% of R1b3 (R-M269) listed are:
Algerians (10.8), Iraquis(10.80),Turks(14.7), Greeks(17.4), Albanians(18.5), Sicilians(24.7), Sardinians(19.2), Spanish(65.6), Portuguese(58.4), Sao Tome(10.1).
AND Tsumkwe Namibian Ju’hoanse San= 6%
Berniell-Lee, G. et al. “Genetic and Demographic Implications of the Bantu Expansion: Insights from Human Paternal Lineages,” Mol. Biol. Evol. 26(7): 1581–1589.
Wood, E. T., et al. 2005 “Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes,” European Journal of Human Genetics 13: 867–876
Hirbo, J.H. 2011 Complex Genetic History of East African Human Populations PhD Dissertation, Department of Biology, University of Maryland
Stop trying to steal the history of Black and African people.
.
quote:Myres, N.M., et al. 2011 “A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe,” European Journal of Human Genetics 19: 95–101
According to the phylogeography of macro-haplogroup K-M9 which contains haplogroup R1b), an ancient Asia-to-Africa back migration has been hypothesized to explain the puzzling presence of R-P25* in sub-Saharan Africa.18 This hypothesis is strongly supported by the present data. In the revised Y chromosome phylogeny, there are 119 lineages in the macro-haplogroup K-M9 (which includes haplogroups K1-K4 and L to T).31 Of these lineages, only two have been observed in sub-Saharan Africa at appreciable frequencies: T-M7018,41,42 and R-V88 (this study). Both haplogroups have also been observed in Europe and western Asia (Refs 42,43 and this study). If the presence of R1b chromosomes in Africa was not because of a back migration, we would have to assume that all the mutations that connect M9 with V88 in the MSY phylogeny (450 mutations) originated in Africa. Under this scenario, we should assume that all the K-M9 lineages that are now found outside sub-Saharan Africa have survived extinction, whereas those which should have accumulated in Africa are now extinct (with the exception of T-M70 and R-V88) and this is an unlikely scenario.
quote:Hirbo p299
The phylogenetic relationships of numerous branches within the core Y-chromosome haplogroup R-M207[haplo R] support a West Asian origin of haplogroup R1b[R1 M173-> M343 R1b], its initial differentiation there followed by a rapid spread of one of its sub-clades carrying the M269 mutation to Europe.
quote:
On the other hand, the majority of European R1b chromosomes carry the marker M269, which is absent in most Africa populations that belong to R1b haplotype. In fact a recent study [431] showed that the geographical distribution of R1b1b2 (R-M269) microsatellite diversity is best explained by spread from a single source in the Near East via Anatolia due to the Neolithic expansion [431].
A recent study of Y chromosome lineages in western and southern Cameroonian populations showed low to moderate frequencies of R1b* that extend into Gabon [294]. Moreover, a recent fine scale sequencing/genotyping of terminal markers in R1b haplotype observed R-V88 sub-haplotype in virtually all the Africans that belong to R1b haplotype [573]. The current study confirms the results of previous work [38, 294] that demonstrates that the R1b* haplotype (meaning R-V88 sub-haplotype) is at its highest frequency in Chadic speaking northern Cameroonian populations (Figure 3.3.2, Appendix 6a). However, the haplotype was not found in any population in East Africa (Figure 3.3.2, Appendix 6a).
quote:This has nothing to do with my Table on M269 in Africa.
Originally posted by Quetzalcoatl:
quote:Can you read? The table purports to show the presence of R-M269 in Africa. I just quoted your sources that there is NO, NONE, ZERO R-M269 in Africa (except a little in Khoisan, and North Africa). You, as usual, did not quote your sources accurately. What I show is that the sources were talking about a different haplotype R-V88 (also named R1b1*) and that even there your numbers were not accurate.
Originally posted by Clyde Winters:
quote:LOL. You admit that the figures in the Table are recorded accurately. And then cite other figures that have nothing to do with M269.
Originally posted by Quetzalcoatl:
quote:Apart from the misleading 53% on Equatorial Guinea there are other problems with the figures you cite. The entire table is titled “African Carriers of R-M269” but in reality R-M269 is extremely rare in Africa. Instead what you are listing are amounts of R1b* (also called R-P25*, R-M343, R-V88). A part of the problem is the unsettled state of the nomenclature and the fact that over the last 10 years different papers have used different names.
Originally posted by Clyde Winters:
.

.
For example in Berniell-Lee:
Your table says R-M269 2-20% in Bantu and in Pygmies 5%, but on
p. 1583 what is listed as present is haplotype R-P25 (R1b1*) with 14% frequency in the Bantu. However, there is zero haplotype R-M269 in either Bantu, or Pygmies
In Wood:
You say R-M269 is present in 2.2 % of the Khoisan. On p. 872 she saysquote:But in Appendix A What we see is a high frequency of R-P25 (R1b1*) in Cameroon But there is ZERO R-M269.
While African haplogroup R chromosomes are generally quite rare, R-P25* chromosomes are found at remarkably high frequencies in northern Cameroon (60.7–94.7%).
Your cite of Hirbo at 6% R-M269 in the Khoisan is correct.
p. 199 Appendix 6a (ii) part III: Frequency of Y chromosome lineages in Africa, Mediterranean and the Near East
what is listed at a very low level are R1b*[R-V88]and R1b3 (Really R-M269)
The frequencies above 10% of R1b3 (R-M269) listed are:
Algerians (10.8), Iraquis(10.80),Turks(14.7), Greeks(17.4), Albanians(18.5), Sicilians(24.7), Sardinians(19.2), Spanish(65.6), Portuguese(58.4), Sao Tome(10.1).
AND Tsumkwe Namibian Ju’hoanse San= 6%
Berniell-Lee, G. et al. “Genetic and Demographic Implications of the Bantu Expansion: Insights from Human Paternal Lineages,” Mol. Biol. Evol. 26(7): 1581–1589.
Wood, E. T., et al. 2005 “Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes,” European Journal of Human Genetics 13: 867–876
Hirbo, J.H. 2011 Complex Genetic History of East African Human Populations PhD Dissertation, Department of Biology, University of Maryland
Stop trying to steal the history of Black and African people.
.
More importantly-both R1b1 and R-M269 are NOT African, but a back flow from Eurasia. Again quoting your sources, which you , as usual, ignore.
Cruciani, F., et al. 2010 “Human Y chromosome haplogroup R-V88: a paternal genetic record of early mid Holocene trans-Saharan
connections and the spread of Chadic languages,” European Journal of Human Genetics 18:800-807
quote:Myres, N.M., et al. 2011 “A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe,” European Journal of Human Genetics 19: 95–101
According to the phylogeography of macro-haplogroup K-M9 which contains haplogroup R1b), an ancient Asia-to-Africa back migration has been hypothesized to explain the puzzling presence of R-P25* in sub-Saharan Africa.18 This hypothesis is strongly supported by the present data. In the revised Y chromosome phylogeny, there are 119 lineages in the macro-haplogroup K-M9 (which includes haplogroups K1-K4 and L to T).31 Of these lineages, only two have been observed in sub-Saharan Africa at appreciable frequencies: T-M7018,41,42 and R-V88 (this study). Both haplogroups have also been observed in Europe and western Asia (Refs 42,43 and this study). If the presence of R1b chromosomes in Africa was not because of a back migration, we would have to assume that all the mutations that connect M9 with V88 in the MSY phylogeny (450 mutations) originated in Africa. Under this scenario, we should assume that all the K-M9 lineages that are now found outside sub-Saharan Africa have survived extinction, whereas those which should have accumulated in Africa are now extinct (with the exception of T-M70 and R-V88) and this is an unlikely scenario.
quote:Hirbo p299
The phylogenetic relationships of numerous branches within the core Y-chromosome haplogroup R-M207[haplo R] support a West Asian origin of haplogroup R1b[R1 M173-> M343 R1b], its initial differentiation there followed by a rapid spread of one of its sub-clades carrying the M269 mutation to Europe.
quote:
On the other hand, the majority of European R1b chromosomes carry the marker M269, which is absent in most Africa populations that belong to R1b haplotype. In fact a recent study [431] showed that the geographical distribution of R1b1b2 (R-M269) microsatellite diversity is best explained by spread from a single source in the Near East via Anatolia due to the Neolithic expansion [431].
A recent study of Y chromosome lineages in western and southern Cameroonian populations showed low to moderate frequencies of R1b* that extend into Gabon [294]. Moreover, a recent fine scale sequencing/genotyping of terminal markers in R1b haplotype observed R-V88 sub-haplotype in virtually all the Africans that belong to R1b haplotype [573]. The current study confirms the results of previous work [38, 294] that demonstrates that the R1b* haplotype (meaning R-V88 sub-haplotype) is at its highest frequency in Chadic speaking northern Cameroonian populations (Figure 3.3.2, Appendix 6a). However, the haplotype was not found in any population in East Africa (Figure 3.3.2, Appendix 6a).

quote:You can whirl , you can spin, you can emit lots of spam BUT there is no R-M269 in Africa, R-V88 (R1b1) is only present in a limited area in Africa, there is no R-M173 (R1) in Africa.
Originally posted by Clyde Winters:
In the Table I cite the source and the frequency. Only a fool and moron would accept your statements as valid given the fact you have repeatedly shown that the figures are correct.
Stop trying to steal the history of Black people.
quote:LOL. If M269 is found in Central, South,North Africa and etc., it is widespread because it is not situated in just one part of the African Continent.
Originally posted by Quetzalcoatl:
quote:You can whirl , you can spin, you can emit lots of spam BUT there is no R-M269 in Africa, R-V88 (R1b1) is only present in a limited area in Africa, there is no R-M173 (R1) in Africa.
Originally posted by Clyde Winters:
In the Table I cite the source and the frequency. Only a fool and moron would accept your statements as valid given the fact you have repeatedly shown that the figures are correct.
Stop trying to steal the history of Black people.
There is nothing to refute. Something has to proven before it can be refuted. It is up to the proposer to produce the evidence for a proposal. You have only produced misquotes. Please quote word-for-word refereed papers that say "M-269 is widespread in Africa" "M-173 is widespread in Africa, "R=P25*(R1b1) is widespread in Africa.


quote:Repeating a lie over and over will not magically turn it into a valid statement. Anyone, including your acolytes (if they are not mesmerized), can go to the papers in question-- all available free and try to find M269 . It is not there. Provide us with a quote-- not a paraphrase or interpretation- that says "R1-M269 is widespread in Africa."
Originally posted by Clyde Winters:
[]LOL. If M269 is found in Central, South,North Africa and etc., it is widespread because it is not situated in just one part of the African Continent.
This supports my contention that this haplogroup is widespread in Africa.

quote:.
Originally posted by Quetzalcoatl:
quote:Repeating a lie over and over will not magically turn it into a valid statement. Anyone, including your acolytes (if they are not mesmerized), can go to the papers in question-- all available free and try to find M269 . It is not there. Provide us with a quote-- not a paraphrase or interpretation- that says "R1-M269 is widespread in Africa."
Originally posted by Clyde Winters:
[]LOL. If M269 is found in Central, South,North Africa and etc., it is widespread because it is not situated in just one part of the African Continent.
This supports my contention that this haplogroup is widespread in Africa.

quote:You are wrong Lee does show that he found M269, eventhough he didn't record the carriers on the Table of Y-chromosomes. .
Originally posted by the lioness,:
quote:this chart made by Clyde is incorrect
Originally posted by Clyde Winters:

example, Biernell-Lee's 2009 paper >>
quote:
Genetic and Demographic Implications of the Bantu Expansion: Insights from Human Paternal Lineages
Biernell-Lee et al 2009
On the whole, most of the samples belonged to previously described African lineages especially common in sub-Saharan Africa. Specifically, most of these lineages have been associated either with Bantu-speaking people—E1b1a (E3a according to The Y Chromosome Consortium 2002), B2a, and E2—or to Pygmy populations (haplogroup B2b). We also observed traces of haplogroups A, E*, E1a, and E1b1b1a (E3b1 according to The Y Chromosome Consortium 2002), which are found at low frequencies across the African continent (Underhill et al. 2000, 2001; Cruciani et al. 2002; Wood et al. 2005). Interestingly, almost 5% of the individuals here analyzed belonged to Eurasian haplogroup R1b1*.
Haplogroup E1b1a, previously proposed as being a marker of the Bantu expansion (Passarino et al. 1998; Scozzari et al. 1999; Underhill et al. 2001; Wood et al. 2005), was the most frequent haplogroup in our sample set (76%), reaching a frequency of almost 80% in agriculturalists and 28% in Pygmy samples (fig. 1).
http://mbe.oxfordjournals.org/content/26/7/1581/F1.large.jpg
As we see here Biernell-Lee reports 0% M269 in the right column of the chart. Insiead R1b1* aka R-P25
And note the numbers are not frequencies. They are just the number of individuals in each group that carried the haplogroup
For instance look again at the quote on E1b1>
E1b1a, previously proposed as being a marker of the Bantu expansion , was the most frequent haplogroup in our sample set (76%), reaching a frequency of almost 80% in agriculturalists and 28% in Pygmy samples
^^^ 76% of the individuals carried E1b1a
But how much on average did those that carried E1b1a ?
Was it 2% or 92%
In this case 80%
That is the frequency, a separate issue as stated
Now let's look at Cruciani 2010 as regards M269 in Africa
Here the figures ARE frequency >
on the far right, haplogroup M269
highest frequency perceatage 2.4% Semitic Egyptians from Baharia
by contrast at the bottom of the chart Western Europeans 57.8%
Perhaps Clyde has R-V69 confused with R-M269 (?)
Clyde Winters is misinterpreting information and making his own charts based on it

quote:This entire paper lacks any merit. I have never said the Dravidians people were Mande speakers. If you read any of my papers you will find that I said
Originally posted by Quetzalcoatl:
For an additional example of Winters's misquoting of sources and refusing to acknowledge that haplotype names change see
https://www.academia.edu/10133899/Clyde_Winters_claim_that_haplotype_M1_proves_that_Indian_Dravidians_are_Mande_speakers_from_Africa_is_not_valid

quote:LOL. Montellano the Kivisild articles makes your whole paper invalid and your arguments prima facie evidence of your lack of knowledge about Indian genetics. In the 1999 article, Kivisild admits that Dravidians carry M1, which is an African haplogroup.
Originally posted by Quetzalcoatl:
For an additional example of Winters's misquoting of sources and refusing to acknowledge that haplotype names change see
https://www.academia.edu/10133899/Clyde_Winters_claim_that_haplotype_M1_proves_that_Indian_Dravidians_are_Mande_speakers_from_Africa_is_not_valid

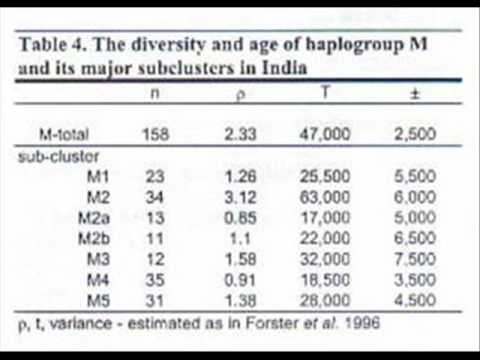
quote:In fact it's the opposite.
Originally posted by the lioness,:
quote:correct, according to what is currently known Haplogroup R did not originate in Africa
Originally posted by Clyde Winters:
The earliest evidence of the R haplogroup comes from Mal’ta Siberia.
quote:
Originally posted by Quetzalcoatl:
quote:You can whirl , you can spin, you can emit lots of spam BUT there is no R-M269 in Africa, R-V88 (R1b1) is only present in a limited area in Africa, there is no R-M173 (R1) in Africa.
Originally posted by Clyde Winters:
In the Table I cite the source and the frequency. Only a fool and moron would accept your statements as valid given the fact you have repeatedly shown that the figures are correct.
Stop trying to steal the history of Black people.
There is nothing to refute. Something has to proven before it can be refuted. It is up to the proposer to produce the evidence for a proposal. You have only produced misquotes. Please quote word-for-word refereed papers that say "M-269 is widespread in Africa" "M-173 is widespread in Africa, "R=P25*(R1b1) is widespread in Africa.
quote:--Hong Shi et al. 2008:
The Y chromosome Alu polymorphism (YAP, also called M1) defines the deep-rooted haplogroup D/E of the global Y-chromosome phylogeny [1]. This D/E haplogroup is further branched into three sub-haplogroups DE*, D and E (Figure 1). The distribution of the D/E haplogroup is highly regional, and the three subgroups are geographically restricted to certain areas, therefore informative in tracing human prehistory (Table 1). The sub-haplogroup DE*, presumably the most ancient lineage of the D/E haplogroup was only found in Africans from Nigeria [2], supporting the "Out of Africa" hypothesis about modern human origin. The sub-haplogroup E (E-M40), defined by M40/SRY4064 and M96, was also suggested originated in Africa [3-6], and later dispersed to Middle East and Europe about 20,000 years ago [3,4]. Interestingly, the sub-haplogroup D defined by M174 (D-M174) is East Asian specific with abundant appearance in Tibetan and Japanese (30–40%), but rare in most of other East Asian populations and populations from regions bordering East Asia (Central Asia, North Asia and Middle East) (usually less than 5%) [5-7]. Under D-M174, Japanese belongs to a separate sub-lineage defined by several mutations (e.g. M55, M57 and M64 etc.), which is different from those in Tibetans implicating relatively deep divergence between them [1]. The fragmented distribution of D-M174 in East Asia seems not consistent with the pattern of other East Asian specific lineages, i.e. O3-M122, O1-M119 and O2-M95 under haplogroup O [8,9].
quote:Haplogroup DE* in Guinea-Bissau:
Further refinement awaits the finding of new markers especially within paragroup E3a*-M2. The microsatellite profile of the DE* individual is one mutational step away from the allelic state described for Nigerians (DYS390*21, DYS388 not tested; [37], therefore suggesting a common ancestry but not elucidating the phylogenetics.
quote:Haplogroup DE* in Nigerians:
There has been considerable debate on the geographic origin of the human Y chromosome Alu polymor- phism (YAP). Here we report a new, very rare deep-rooting haplogroup within the YAP clade, together with data on other deep-rooting YAP clades. The new haplogroup, found so far in only five Nigerians, is the least-derived YAP haplogroup according to currently known binary markers. However, because the interior branching order of the Y chromosome genealogical tree remains unknown, it is impossible to impute the origin of the YAP clade with certainty. We discuss the problems presented by rare deep-rooting lineages for Y chromosome phylogeography.
quote:--Fulvio Cruciani et al.
‘‘Out of Africa’’ haplogroups.
All Y-clades that are not exclusively African belong to the macro-haplogroup CT, which is defined by mutations M168, M294 and P9.1 [14,31] and is subdivided into two major clades, DE and CF [1,14].
In a recent study [16], sequencing of two chromosomes belonging to haplogroups C and R, led to the identification of 25 new mutations, eleven of which were in the C-chromosome and seven in the R-chromosome.
Here, the seven mutations which were found to be shared by chromosomes of haplogroups C and R [16], were also found to be present in one DE sample (sample 33 in Table S1), and positioned at the root of macro-haplogroup CT (Figure 1 and Figure S1)
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3492319/figure/pone-0049170-g001/
Figure S1
Structure of the macro-haplogroup CT. For details on mutations see legend to Figure 1. Dashed lines indicate putative branchings (no positive control available). The position of V248 (haplogroup C2) and V87 (haplogroup C3) compared to mutations that define internal branches was not determined. Note that mutations V45, V69 and V88 have been previously mapped (Cruciani et al. 2010; Eur J Hum Genet 18∶800–807).
(TIF)
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3492319/bin/pone.0049170.s001.tif
quote:Use of Y Chromosome and Mitochondrial DNA Population Structure in Tracing Human Migrations
"haplogroup CF and DE molecular ancestors first evolved inside Africa and subsequently contributed as Y chromosome founders to pioneering migrations that successfully colonized Asia. While not proof, the DE and CF bifurcation (Figure 8d ) is consistent with independent colonization impulses possibly occurring in a short time interval."
quote:Basal!
Originally posted by the lioness,:
Not all haplogroups originate in Africa even if they're are also found in Africa because it depends on where they started.
The origin of a haplogroup is strongly suggested by
a) high frequency
b) high diversity
c) DNA analysis of ancient human remains

quote:LOL. Montellano the Kivisild articles makes your whole paper invalid and your arguments prima facie evidence of your lack of knowledge about Indian genetics. In the 1999 article, Kivisild admits that Dravidians carry M1, which is an African haplogroup.
Originally posted by Quetzalcoatl:
For an additional example of Winters's misquoting of sources and refusing to acknowledge that haplotype names change see
https://www.academia.edu/10133899/Clyde_Winters_claim_that_haplotype_M1_proves_that_Indian_Dravidians_are_Mande_speakers_from_Africa_is_not_valid


quote:Correct. Geneticists like to look at haplogroups in isolation. The archaeology makes it clear that the first Europeans came from Africa and they were Khoisan, because the same culutral traditions of Cro-Magnon man in Europe were found in Southern Africa. It is clear that Mal'ta man's population did not just fall from the sky.
Originally posted by Troll Patrol # Ish Gebor:
quote:
Originally posted by Quetzalcoatl:
quote:You can whirl , you can spin, you can emit lots of spam BUT there is no R-M269 in Africa, R-V88 (R1b1) is only present in a limited area in Africa, there is no R-M173 (R1) in Africa.
Originally posted by Clyde Winters:
In the Table I cite the source and the frequency. Only a fool and moron would accept your statements as valid given the fact you have repeatedly shown that the figures are correct.
Stop trying to steal the history of Black people.
There is nothing to refute. Something has to proven before it can be refuted. It is up to the proposer to produce the evidence for a proposal. You have only produced misquotes. Please quote word-for-word refereed papers that say "M-269 is widespread in Africa" "M-173 is widespread in Africa, "R=P25*(R1b1) is widespread in Africa.quote:--Hong Shi et al. 2008:
The Y chromosome Alu polymorphism (YAP, also called M1) defines the deep-rooted haplogroup D/E of the global Y-chromosome phylogeny [1]. This D/E haplogroup is further branched into three sub-haplogroups DE*, D and E (Figure 1). The distribution of the D/E haplogroup is highly regional, and the three subgroups are geographically restricted to certain areas, therefore informative in tracing human prehistory (Table 1). The sub-haplogroup DE*, presumably the most ancient lineage of the D/E haplogroup was only found in Africans from Nigeria [2], supporting the "Out of Africa" hypothesis about modern human origin. The sub-haplogroup E (E-M40), defined by M40/SRY4064 and M96, was also suggested originated in Africa [3-6], and later dispersed to Middle East and Europe about 20,000 years ago [3,4]. Interestingly, the sub-haplogroup D defined by M174 (D-M174) is East Asian specific with abundant appearance in Tibetan and Japanese (30–40%), but rare in most of other East Asian populations and populations from regions bordering East Asia (Central Asia, North Asia and Middle East) (usually less than 5%) [5-7]. Under D-M174, Japanese belongs to a separate sub-lineage defined by several mutations (e.g. M55, M57 and M64 etc.), which is different from those in Tibetans implicating relatively deep divergence between them [1]. The fragmented distribution of D-M174 in East Asia seems not consistent with the pattern of other East Asian specific lineages, i.e. O3-M122, O1-M119 and O2-M95 under haplogroup O [8,9].
http://www.biomedcentral.com/1741-7007/6/45
quote:Haplogroup DE* in Guinea-Bissau:
Further refinement awaits the finding of new markers especially within paragroup E3a*-M2. The microsatellite profile of the DE* individual is one mutational step away from the allelic state described for Nigerians (DYS390*21, DYS388 not tested; [37], therefore suggesting a common ancestry but not elucidating the phylogenetics.
Y-chromosomal diversity in the population of Guinea-Bissau: a multiethnic perspective
http://www.biomedcentral.com/1471-2148/7/124
quote:Haplogroup DE* in Nigerians:
There has been considerable debate on the geographic origin of the human Y chromosome Alu polymor- phism (YAP). Here we report a new, very rare deep-rooting haplogroup within the YAP clade, together with data on other deep-rooting YAP clades. The new haplogroup, found so far in only five Nigerians, is the least-derived YAP haplogroup according to currently known binary markers. However, because the interior branching order of the Y chromosome genealogical tree remains unknown, it is impossible to impute the origin of the YAP clade with certainty. We discuss the problems presented by rare deep-rooting lineages for Y chromosome phylogeography.
Rare Deep-Rooting Y Chromosome Lineages in Humans: Lessons for Phylogeography
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1462739/pdf/14504230.pdf
Y-DNA haplogroup R-M207 is believed to have arisen approximately 27,000 years ago in Asia. The two currently defined subclades are R1 and R2.
quote:--Fulvio Cruciani et al.
‘‘Out of Africa’’ haplogroups.
All Y-clades that are not exclusively African belong to the macro-haplogroup CT, which is defined by mutations M168, M294 and P9.1 [14,31] and is subdivided into two major clades, DE and CF [1,14].
In a recent study [16], sequencing of two chromosomes belonging to haplogroups C and R, led to the identification of 25 new mutations, eleven of which were in the C-chromosome and seven in the R-chromosome.
Here, the seven mutations which were found to be shared by chromosomes of haplogroups C and R [16], were also found to be present in one DE sample (sample 33 in Table S1), and positioned at the root of macro-haplogroup CT (Figure 1 and Figure S1)
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3492319/figure/pone-0049170-g001/
Figure S1
Structure of the macro-haplogroup CT. For details on mutations see legend to Figure 1. Dashed lines indicate putative branchings (no positive control available). The position of V248 (haplogroup C2) and V87 (haplogroup C3) compared to mutations that define internal branches was not determined. Note that mutations V45, V69 and V88 have been previously mapped (Cruciani et al. 2010; Eur J Hum Genet 18∶800–807).
(TIF)
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3492319/bin/pone.0049170.s001.tif
Molecular Dissection of the Basal Clades in the Human Y Chromosome Phylogenetic Tree (2011)
quote:Use of Y Chromosome and Mitochondrial DNA Population Structure in Tracing Human Migrations
"haplogroup CF and DE molecular ancestors first evolved inside Africa and subsequently contributed as Y chromosome founders to pioneering migrations that successfully colonized Asia. While not proof, the DE and CF bifurcation (Figure 8d ) is consistent with independent colonization impulses possibly occurring in a short time interval."
--Peter A. Underhill , Toomas Kivisild - 2007
The Mal'ta boy didn't fell from the sky, onto Siberia near Lake Baikal?
quote:High frequency and diversity can not tell us anything about origin because these features are are associated with contemporary populations. Since they relate only to contemporary populations, the story they tell is about the people who presently live in the area. These markers have little validity, because the contemporary population may not represent the original inhabitant of a particular region. This is especially true of European whites who only recently settled Europe, after they migrated from Central Asia.
Originally posted by the lioness,:
Not all haplogroups originate in Africa even if they're are also found in Africa because it depends on where they started.
The origin of a haplogroup is strongly suggested by
a) high frequency
b) high diversity
c) DNA analysis of ancient human remains
quote:As you can see I did admit that only 5.2% of the pygmy carried R1.
Y-chromosome R1 is found throghout Africa. The pristine
form of R1-M173 is only found in Africa (Coia et al,
2005; Cruciani et al, 2002, 2010). The age of ychromosome
R is 27ky. Most researchers believe that
R(M173) is 18.5 ky.There is a great diversity of the
macrohaplogroup R in Africa (See Figure 1). Ychromosome
R is characterized by M207/V45. The V45
mutation is found among African populations ( Cruciani et
al ,2010). ISOGG 2010 Y-DNA haplogroup tree makes it
clear that V45 is phylogenetically equivalent to M207.The
most common R haplogroup in Africa is R1 (M173). The
predominant haplogroup is R1b (Berniell-Lee et al,2009;
Coia et al, 2005; Winters, 2010b; Wood et al, 2009).
Cruciani et al (2010) discovered new R1b mutations
including V7, V8, V45, V69, and V88. Geography appears
to play a significant role in the distribution of haplogroup
R in Africa. Cruciani et al (2010) has renamed the R*-
M173 (R P-25) in Africa V88. The TMRCA of V88 was
9200-5600 kya (Cruciani et al, 2010).
Y-chromosome V88 (R1b1a) has its highest frequency
among Chadic speakers, while the carriers of V88 among
Niger-Congo speakers (predominately Bantu people)
range between 2-66% ( Cruciani et al, 2010; Bernielle-Lee
et al, 2009). Haplogroup V88 includes the mutations M18,
V35 and V7. Cruciani et al (2010) revealed that R-V88 is
also carried by Eurasians including the distinctive
mutations M18, V35 and V7.
R1b1-P25 is found in Western Eurasia. Haplogroup
R1b1* is found in Africa at various frequencies. BerniellLee
et al (2009) found in their study that 5.2% carried
Rb1*. The frequency of R1b1* among the Bantu ranged
from 2-20. The bearers of R1b1* among the Pygmy
populations ranged from 1-25% (Berniell-Lee et al, 2009).
The frequency of R1b1 among Guinea-Bissau populations
was 12% (Carvalho et al,2010).
quote:As a result, I did not attempt to decieve anyone about the frequency of R1 in Africa as the author implies.
Around 0.1 of Sub Saharan Africans carry R1b1b2. Wood
et al (2009) found that Khoisan (2.2%) and Niger-Congo
(0.4%) speakers carried the R-M269 y-chromosome.
The Khoisan also carry RM343 (R1b) and M 198 (R1a1)
(Naidoo et al., 2010) the archaeological and linguistic data
indicate the successful colonization of Asia by SubSaharan
Africans from Nubia 5-4kya (Winters, 2007,2008,
2010c). The archaeological evidence makes it clear that
around 4kya intercultural style artifacts connected Africa
and Eurasia (Winters, 2007,2010c).




quote:Predictably, the air wavs are now filled with your irrelevant spam.
Originally posted by Clyde Winters:
quote:LOL. Montellano the Kivisild articles makes your whole paper invalid and your arguments prima facie evidence of your lack of knowledge about Indian genetics. In the 1999 article, Kivisild admits that Dravidians carry M1, which is an African haplogroup.
Originally posted by Quetzalcoatl:
For an additional example of Winters's misquoting of sources and refusing to acknowledge that haplotype names change see
https://www.academia.edu/10133899/Clyde_Winters_claim_that_haplotype_M1_proves_that_Indian_Dravidians_are_Mande_speakers_from_Africa_is_not_valid
This article is just as relevant today as it was in 1999. People carry a particular gene do not just disappear.
The Eastern African hg M1, HVS-I signature motif is 16,129, 16,189, 16,223, 16,249, and 16,311. In the Kivisild et al figure below we see the same motif. The mutations are shown less 16,000.
Here you can clearly see:mutations 129,189, 223 and 311, in Indian M1, in Figure 3, of Kivisild et al, 1999.

In the Kivisild et al 1999 study of Indian mtDNA around 15% carried haplogroup M1. See:
http://evolutsioon.ut.ee/publications/Kivisild1999b.pdf
.

In Table 4, Kivisild et al, 1999, we see the frequency of M1 in India.There are 217 million Dravidian speakers in India, if we compare the frequency of M1 carriers to the Dravidian speaking community around 32 million people carry M1.
The frequency of 15% of the Dravidians carrying M1 shows the presence of M1 in India. As a result, your arguments are false and invalid.
quote:.
Originally posted by Clyde Winters:
[LOL. Montellano the Kivisild articles makes your whole paper invalid and your arguments prima facie evidence of your lack of knowledge about Indian genetics. In the 1999 article, Kivisild admits that Dravidians carry M1, which is an African haplogroup.
This article is just as relevant today as it was in 1999. People carry a particular gene do not just disappear.
quote:If anyone wants to check- look up SNPs 129, 189,223, 249 and 311 in http://www.phylotree.org/Y/tree/index.htm and see that the name is M3 not M1
The Eastern African hg M1, HVS-I signature motif is 16,129, 16,189, 16,223, 16,249, and 16,311. In the Kivisild et al figure below we see the same motif. The mutations are shown less 16,000.
quote:NO, 23 million people in India carry M3. Get over it!!
In Table 4, Kivisild et al, 1999, we see the frequency of M1 in India.There are 217 million Dravidian speakers in India, if we compare the frequency of M1 carriers to the Dravidian speaking community around 32 million people carry M1. [QB][/
The frequency of 15% of the Dravidians carrying M1 shows the presence of M1 in India. As a result, your arguments are false and invalid.
quote:Yep,
Originally posted by Clyde Winters:
quote:Correct. Geneticists like to look at haplogroups in isolation. The archaeology makes it clear that the first Europeans came from Africa and they were Khoisan, because the same culutral traditions of Cro-Magnon man in Europe were found in Southern Africa. It is clear that Mal'ta man's population did not just fall from the sky.
Originally posted by Troll Patrol # Ish Gebor:
quote:
Originally posted by Quetzalcoatl:
quote:You can whirl , you can spin, you can emit lots of spam BUT there is no R-M269 in Africa, R-V88 (R1b1) is only present in a limited area in Africa, there is no R-M173 (R1) in Africa.
Originally posted by Clyde Winters:
In the Table I cite the source and the frequency. Only a fool and moron would accept your statements as valid given the fact you have repeatedly shown that the figures are correct.
Stop trying to steal the history of Black people.
There is nothing to refute. Something has to proven before it can be refuted. It is up to the proposer to produce the evidence for a proposal. You have only produced misquotes. Please quote word-for-word refereed papers that say "M-269 is widespread in Africa" "M-173 is widespread in Africa, "R=P25*(R1b1) is widespread in Africa.quote:--Hong Shi et al. 2008:
The Y chromosome Alu polymorphism (YAP, also called M1) defines the deep-rooted haplogroup D/E of the global Y-chromosome phylogeny [1]. This D/E haplogroup is further branched into three sub-haplogroups DE*, D and E (Figure 1). The distribution of the D/E haplogroup is highly regional, and the three subgroups are geographically restricted to certain areas, therefore informative in tracing human prehistory (Table 1). The sub-haplogroup DE*, presumably the most ancient lineage of the D/E haplogroup was only found in Africans from Nigeria [2], supporting the "Out of Africa" hypothesis about modern human origin. The sub-haplogroup E (E-M40), defined by M40/SRY4064 and M96, was also suggested originated in Africa [3-6], and later dispersed to Middle East and Europe about 20,000 years ago [3,4]. Interestingly, the sub-haplogroup D defined by M174 (D-M174) is East Asian specific with abundant appearance in Tibetan and Japanese (30–40%), but rare in most of other East Asian populations and populations from regions bordering East Asia (Central Asia, North Asia and Middle East) (usually less than 5%) [5-7]. Under D-M174, Japanese belongs to a separate sub-lineage defined by several mutations (e.g. M55, M57 and M64 etc.), which is different from those in Tibetans implicating relatively deep divergence between them [1]. The fragmented distribution of D-M174 in East Asia seems not consistent with the pattern of other East Asian specific lineages, i.e. O3-M122, O1-M119 and O2-M95 under haplogroup O [8,9].
http://www.biomedcentral.com/1741-7007/6/45
quote:Haplogroup DE* in Guinea-Bissau:
Further refinement awaits the finding of new markers especially within paragroup E3a*-M2. The microsatellite profile of the DE* individual is one mutational step away from the allelic state described for Nigerians (DYS390*21, DYS388 not tested; [37], therefore suggesting a common ancestry but not elucidating the phylogenetics.
Y-chromosomal diversity in the population of Guinea-Bissau: a multiethnic perspective
http://www.biomedcentral.com/1471-2148/7/124
quote:Haplogroup DE* in Nigerians:
There has been considerable debate on the geographic origin of the human Y chromosome Alu polymor- phism (YAP). Here we report a new, very rare deep-rooting haplogroup within the YAP clade, together with data on other deep-rooting YAP clades. The new haplogroup, found so far in only five Nigerians, is the least-derived YAP haplogroup according to currently known binary markers. However, because the interior branching order of the Y chromosome genealogical tree remains unknown, it is impossible to impute the origin of the YAP clade with certainty. We discuss the problems presented by rare deep-rooting lineages for Y chromosome phylogeography.
Rare Deep-Rooting Y Chromosome Lineages in Humans: Lessons for Phylogeography
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1462739/pdf/14504230.pdf
Y-DNA haplogroup R-M207 is believed to have arisen approximately 27,000 years ago in Asia. The two currently defined subclades are R1 and R2.
quote:--Fulvio Cruciani et al.
‘‘Out of Africa’’ haplogroups.
All Y-clades that are not exclusively African belong to the macro-haplogroup CT, which is defined by mutations M168, M294 and P9.1 [14,31] and is subdivided into two major clades, DE and CF [1,14].
In a recent study [16], sequencing of two chromosomes belonging to haplogroups C and R, led to the identification of 25 new mutations, eleven of which were in the C-chromosome and seven in the R-chromosome.
Here, the seven mutations which were found to be shared by chromosomes of haplogroups C and R [16], were also found to be present in one DE sample (sample 33 in Table S1), and positioned at the root of macro-haplogroup CT (Figure 1 and Figure S1)
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3492319/figure/pone-0049170-g001/
Figure S1
Structure of the macro-haplogroup CT. For details on mutations see legend to Figure 1. Dashed lines indicate putative branchings (no positive control available). The position of V248 (haplogroup C2) and V87 (haplogroup C3) compared to mutations that define internal branches was not determined. Note that mutations V45, V69 and V88 have been previously mapped (Cruciani et al. 2010; Eur J Hum Genet 18∶800–807).
(TIF)
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3492319/bin/pone.0049170.s001.tif
Molecular Dissection of the Basal Clades in the Human Y Chromosome Phylogenetic Tree (2011)
quote:Use of Y Chromosome and Mitochondrial DNA Population Structure in Tracing Human Migrations
"haplogroup CF and DE molecular ancestors first evolved inside Africa and subsequently contributed as Y chromosome founders to pioneering migrations that successfully colonized Asia. While not proof, the DE and CF bifurcation (Figure 8d ) is consistent with independent colonization impulses possibly occurring in a short time interval."
--Peter A. Underhill , Toomas Kivisild - 2007
The Mal'ta boy didn't fell from the sky, onto Siberia near Lake Baikal?
quote:--Erwan Pennarun, Toomas Kivisild
"No southwest Asian specific clades for M1 or U6 were discovered. U6 and M1 frequencies in North Africa, the Middle East and Europe DO NOT FOLLOW similar patterns, and their sub-clade divisions do not appear to be compatible with their shared history reaching back to the Early Upper Palaeolithic."
quote:-- Petraglia, M and Rose, J
“... the M1 presence in the Arabian
peninsula signals a predominant East
African influence since the Neolithic
onwards.“
quote:LOL. How could haplogroup M1 change to M3, when Kivisild presents both haplohroups in Figure 3.
Originally posted by Quetzalcoatl:
quote:.
Originally posted by Clyde Winters:
[LOL. Montellano the Kivisild articles makes your whole paper invalid and your arguments prima facie evidence of your lack of knowledge about Indian genetics. In the 1999 article, Kivisild admits that Dravidians carry M1, which is an African haplogroup.
This article is just as relevant today as it was in 1999. People carry a particular gene do not just disappear.
The 1999 paper is invalid. The people did not disappear, however the name of their haplotype changed (by the committee in charge of nomenclature) to M3.
quote:If anyone wants to check- look up SNPs 129, 189,223, 249 and 311 in http://www.phylotree.org/Y/tree/index.htm and see that the name is M3 not M1
The Eastern African hg M1, HVS-I signature motif is 16,129, 16,189, 16,223, 16,249, and 16,311. In the Kivisild et al figure below we see the same motif. The mutations are shown less 16,000.
quote:NO, 23 million people in India carry M3. Get over it!!
In Table 4, Kivisild et al, 1999, we see the frequency of M1 in India.There are 217 million Dravidian speakers in India, if we compare the frequency of M1 carriers to the Dravidian speaking community around 32 million people carry M1. [QB][/
The frequency of 15% of the Dravidians carrying M1 shows the presence of M1 in India. As a result, your arguments are false and invalid.

quote:Abu-Amero's 2009 article is called Saudi Arabian Y-Chromosome diversity and its relationship with with nearby regions.
Originally posted by Clyde Winters:
Abu-Amero et al (2009) reveal the fact that Dravidians carry the R haplogroups illustrate the recent introduction of R y-chromosomes to Eurasia. The frequency of haplotype M173 in Eurasia is as follows: Anatolia 0.19%, Iran 2.67%, Iraq 0.49% Oman 1.0%, Pakistan 0.57% and Oman 1.0% . This contrast sharply with the widespread distribution of R1 in Africa that ranges between 7- 95% in various parts of Africa,
quote:These questions have nothing to do with this thread.
Originally posted by the lioness,:
quote:Abu-Amero's 2009 article is called Saudi Arabian Y-Chromosome diversity and its relationship with with nearby regions.
Originally posted by Clyde Winters:
Abu-Amero et al (2009) reveal the fact that Dravidians carry the R haplogroups illustrate the recent introduction of R y-chromosomes to Eurasia. The frequency of haplotype M173 in Eurasia is as follows: Anatolia 0.19%, Iran 2.67%, Iraq 0.49% Oman 1.0%, Pakistan 0.57% and Oman 1.0% . This contrast sharply with the widespread distribution of R1 in Africa that ranges between 7- 95% in various parts of Africa,
www.ncbi.nlm.nih.gov/pmc/articles/PMC2759955/
The regions discussed in the article are far from being the whole of Eurasia
Saudi Arabia
Qatar
UAE
Oman
Yemen
Somalia
Egypt
Jordan
Iraq
Anataolia
Iran
Pakistan
Lebanon
Why are you saying that the article reveals the fact that Dravidians carry the R haplogroups when the article says nothing about Dravidians ?
Why are you saying the frequency of haplotype M173 in Eurasia is represented by the above countries when the above countries are only a particular region of Eurasia and are NOT the countries with the HIGH frequencies of M173 ?
________________________________
As for M17 the descendant of M173 in India it is found at the higher frequencies amoung the Indo European speakers, the Hindi spekaing Brahmin and at lower frequencies in the Dravidians
And all of these people of course also Eurasians
An it is also found in high frequenices in Central Asians and some Eastern Europeans
You like to speak about the Aryan invasions of India. That of course will also have a genetic marker
We are talking about R1a here
Of course in Cameroon it's a clade of R1b not R1a
quote:Clyde I can't believe you don't know this, haplogroup R-M17 is the sub clade of M173 found in India.
Originally posted by Clyde Winters:
quote:These questions have nothing to do with this thread.
Originally posted by the lioness,:
quote:Abu-Amero's 2009 article is called Saudi Arabian Y-Chromosome diversity and its relationship with with nearby regions.
Originally posted by Clyde Winters:
Abu-Amero et al (2009) reveal the fact that Dravidians carry the R haplogroups illustrate the recent introduction of R y-chromosomes to Eurasia. The frequency of haplotype M173 in Eurasia is as follows: Anatolia 0.19%, Iran 2.67%, Iraq 0.49% Oman 1.0%, Pakistan 0.57% and Oman 1.0% . This contrast sharply with the widespread distribution of R1 in Africa that ranges between 7- 95% in various parts of Africa,
www.ncbi.nlm.nih.gov/pmc/articles/PMC2759955/
The regions discussed in the article are far from being the whole of Eurasia
Saudi Arabia
Qatar
UAE
Oman
Yemen
Somalia
Egypt
Jordan
Iraq
Anataolia
Iran
Pakistan
Lebanon
Why are you saying that the article reveals the fact that Dravidians carry the R haplogroups when the article says nothing about Dravidians ?
Why are you saying the frequency of haplotype M173 in Eurasia is represented by the above countries when the above countries are only a particular region of Eurasia and are NOT the countries with the HIGH frequencies of M173 ?
________________________________
As for M17 the descendant of M173 in India it is found at the higher frequencies amoung the Indo European speakers, the Hindi spekaing Brahmin and at lower frequencies in the Dravidians
And all of these people of course also Eurasians
An it is also found in high frequenices in Central Asians and some Eastern Europeans
You like to speak about the Aryan invasions of India. That of course will also have a genetic marker
We are talking about R1a here
Of course in Cameroon it's a clade of R1b not R1a
We are talking about M173 in Africa and M1 in India.
Stay on topic
.
quote:^^^ So if you are going to make very misleading statements like this don't expect it to go unaddressed
Originally posted by Clyde Winters:
Abu-Amero et al (2009) reveal the fact that Dravidians carry the R haplogroups illustrate the recent introduction of R y-chromosomes to Eurasia. The frequency of haplotype M173 in Eurasia is as follows: Anatolia 0.19%, Iran 2.67%, Iraq 0.49% Oman 1.0%, Pakistan 0.57% and Oman 1.0% . This contrast sharply with the widespread distribution of R1 in Africa that ranges between 7- 95% in various parts of Africa,
quote:--Scozzari R, Massaia A, D’Atanasio E, Myres NM, Perego UA, et al.(2012) Molecular Dissection of the Basal Clades in the Human Y Chromosome Phylogenetic Tree. PLoS ONE 7(11): e49170. doi:10.1371/journal.pone.00491 70
An independent high resolution MSY phylogeny has been recently obtained from 2,870 Y-SNPs discovered (or re-discovered) in the course of a large whole-genome re-sequencing study, but the observed variable sites all belong to the recent “out of Africa” CT clade [15]. Recently, in a re-sequencing study of the Y chromosome, the root of the tree moved to a new position and several changes at the basal nodes of the phylogeny were introduced [16]. Interestingly, the estimated coalescence age and deep branching pattern of the revised MSY tree appear to be more similar to those of the mtDNA phylogeny [17],[18] than previously reported [1].
[...]
Three of the seven R-specific mutations (V45, V69 and V88) were previously mapped within haplogroup R [34], whereas the remaining four mutations have been here positioned at the root of haplogroups F (V186 and V205), K (V104) and P (V231)(Figure S1) through the analysis of 12 haplogroup F samples (samples 40–51, in Table S1).
[...]
All Y-clades that are not exclusively African belong to the macro-haplogroup CT, which is defined by mutations M168, M294 and P9.1 [14],[31] and is subdivided into two major clades, DE and CF [1],[14]. In a recent study [16], sequencing of two chromosomes belonging to haplogroups C and R, led to the identification of 25 new mutations, eleven of which were in the C-chromosome and seven in the R-chromosome. Here, the seven mutations which were found to be shared by chromosomes of haplogroups C and R [16]
[...]
Two A1b chromosomes from a previous work (one from Algeria and one from Cameroon)[16] were included in this study together with two newly identified A1b chromosomes, whose geographic origin can be traced back to west-central Africa (Ghana) on the basis of the microsatellite profile (data not shown).
Structure of the macro-haplogroup CT. For details on mutations see legend to Figure 1. Dashed lines indicate putative branchings (no positive control available). The position of V248 (haplogroup C2) and V87 (haplogroup C3) compared to mutations that define internal branches was not determined. Note that mutations V45, V69 and V88 have been previously mapped (Cruciani et al. 2010; Eur J Hum Genet 18∶800–807).
quote:This is irrelevant because I cite the source for the frequency of haplotype M173 in Eurasia, which are much lower than those in Africa. These figures are not mine.
Originally posted by the lioness,:
quote:^^^ So if you are going to make very misleading statements like this don't expect it to go unaddressed
Originally posted by Clyde Winters:
Abu-Amero et al (2009) reveal the fact that Dravidians carry the R haplogroups illustrate the recent introduction of R y-chromosomes to Eurasia. The frequency of haplotype M173 in Eurasia is as follows: Anatolia 0.19%, Iran 2.67%, Iraq 0.49% Oman 1.0%, Pakistan 0.57% and Oman 1.0% . This contrast sharply with the widespread distribution of R1 in Africa that ranges between 7- 95% in various parts of Africa,




quote:I wrote you about it on September 29 at 1:59 PM on the melanin thread http://www.egyptsearch.com/forums/ultimatebb.cgi?ubb=get_topic;f=15;t=010486 with some other comments
Originally posted by tropicals redacted:
@Quetzalcoatl
How did you get on with Pagani?
The finding that the modern Egyptian sample (100) in his 2015 paper was 80% non-African?
You said you thought of e-mailing him to query it.
quote:You quoted a source on frequencies of R1 in Saudi Arabia and nearby countries that have low frequencies of R1 and said that represented Eurasia
Originally posted by Clyde Winters:
quote:This is irrelevant because I cite the source for the frequency of haplotype M173 in Eurasia, which are much lower than those in Africa. These figures are not mine.
Originally posted by the lioness,:
[qb]
quote:^^^ So if you are going to make very misleading statements like this don't expect it to go unaddressed
Originally posted by Clyde Winters:
Abu-Amero et al (2009) reveal the fact that Dravidians carry the R haplogroups illustrate the recent introduction of R y-chromosomes to Eurasia. The frequency of haplotype M173 in Eurasia is as follows: Anatolia 0.19%, Iran 2.67%, Iraq 0.49% Oman 1.0%, Pakistan 0.57% and Oman 1.0% . This contrast sharply with the widespread distribution of R1 in Africa that ranges between 7- 95% in various parts of Africa,
quote:I was curious to what the outcome.
Originally posted by Quetzalcoatl:
quote:I wrote you about it on September 29 at 1:59 PM on the melanin thread http://www.egyptsearch.com/forums/ultimatebb.cgi?ubb=get_topic;f=15;t=010486 with some other comments
Originally posted by tropicals redacted:
@Quetzalcoatl
How did you get on with Pagani?
The finding that the modern Egyptian sample (100) in his 2015 paper was 80% non-African?
You said you thought of e-mailing him to query it.
quote:Here, the seven mutations which were found to be shared by chromosomes of haplogroups C and R [16], were also found to be present in one DE sample (sample 33 in Table S1), and positioned at the root of macro-haplogroup CT (Figure 1 and Figure S1)
Originally posted by the lioness,:
quote:You quoted a source on frequencies of R1 in Saudi Arabia and nearby countries that have low frequencies of R1 and said that represented Eurasia
Originally posted by Clyde Winters:
quote:This is irrelevant because I cite the source for the frequency of haplotype M173 in Eurasia, which are much lower than those in Africa. These figures are not mine.
Originally posted by the lioness,:
[qb]
quote:^^^ So if you are going to make very misleading statements like this don't expect it to go unaddressed
Originally posted by Clyde Winters:
Abu-Amero et al (2009) reveal the fact that Dravidians carry the R haplogroups illustrate the recent introduction of R y-chromosomes to Eurasia. The frequency of haplotype M173 in Eurasia is as follows: Anatolia 0.19%, Iran 2.67%, Iraq 0.49% Oman 1.0%, Pakistan 0.57% and Oman 1.0% . This contrast sharply with the widespread distribution of R1 in Africa that ranges between 7- 95% in various parts of Africa,
Meanwhile you left out R1 frequencies of Europe, India and Central Asia.
Everybody knows what you did

quote:There is more than one M haplogroup in Africa.
Originally posted by the lioness,:
The idea that haplgroup M maybe have originated in East Africa is a legitiimate theory but may or may not be true since the horn is a coastal region and has higher frequencies of Eurasian haplogroups as compared to other parts of sub saharan Africa
Of the various sub clades only M1 is found in Africa, in the horn. Ethiopians have the highest frequencies at about 20%.
M is the single most common mtDNA haplogroup in Asia and in the world is most diverse in India.
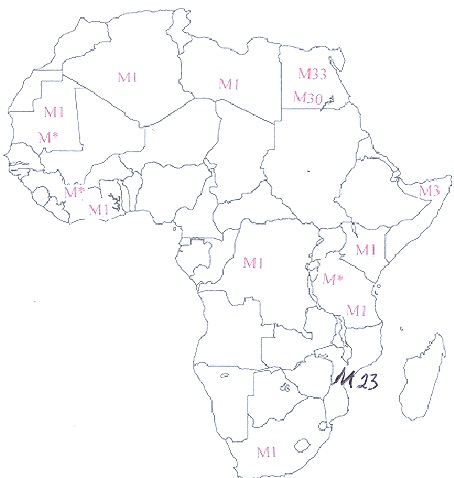
quote:
Originally posted by the lioness,:
The idea that haplgroup M maybe have originated in East Africa is a legitiimate theory but may or may not be true since the horn is a coastal region and has higher frequencies of Eurasian haplogroups as compared to other parts of sub saharan Africa
Of the various sub clades only M1 is found in Africa, in the horn. Ethiopians have the highest frequencies at about 20%.
M is the single most common mtDNA haplogroup in Asia and in the world is most diverse in India.
![[Frown]](frown.gif) Not again.
Not again. quote:--Adimoolam Chandrasekar et al. 2009
"The presence of M haplogroup in Ethiopia, named M1, led to the proposal that haplogroup M originated in eastern Africa, approximately 60,000 years ago, and was carried towards Asia [34].
Macrohaplogroup M is ubiquitous in India and covers more than 70 per cent of the Indian mtDNA lineages [28], [36]–[38]. Recent studies on complete mtDNA sequences (~187) tried to resolve the phylogeny of Indian macrohaplogroup M. As a result, M2, M3, M4, M5, M6 [28], [36], [39]–[40], M18, M25 [38], M30, [41], M31 [42], [24] M33, M34, M35, M36, M37, M38, M39, M40 [22], M41, M42 [43], M43 [23], [44], M45 [45], M48, M49, and M50 [46] haplogroups of M that was identified in India helped to a certain extent in understanding M genealogy in diversified Indian populations. In the above background, extensive sequencing of complete mtDNA of South Asia, particularly India, is essential for better understanding of the peopling of the non-African continents, and pathogenesis of diseases in various ethnic groups with different matrilineal backgrounds."
quote:That fact that a haplogroup is found in Africa doesn't means it originates there. '
Originally posted by Clyde Winters:
quote:There is more than one M haplogroup in Africa.
Originally posted by the lioness,:
The idea that haplgroup M maybe have originated in East Africa is a legitiimate theory but may or may not be true since the horn is a coastal region and has higher frequencies of Eurasian haplogroups as compared to other parts of sub saharan Africa
Of the various sub clades only M1 is found in Africa, in the horn. Ethiopians have the highest frequencies at about 20%.
M is the single most common mtDNA haplogroup in Asia and in the world is most diverse in India.
You can read more African Origin of M1
African Origin hg M23
Enjoy
.
quote:This map shows the diversity of haplogroup M in Africa.
Originally posted by the lioness,:
quote:That fact that a haplogroup is found in Africa doesn't means it originates there. '
Originally posted by Clyde Winters:
quote:There is more than one M haplogroup in Africa.
Originally posted by the lioness,:
The idea that haplgroup M maybe have originated in East Africa is a legitiimate theory but may or may not be true since the horn is a coastal region and has higher frequencies of Eurasian haplogroups as compared to other parts of sub saharan Africa
Of the various sub clades only M1 is found in Africa, in the horn. Ethiopians have the highest frequencies at about 20%.
M is the single most common mtDNA haplogroup in Asia and in the world is most diverse in India.
You can read more African Origin of M1
African Origin hg M23
Enjoy
.
Some haplogroups found in modern Africans are the result of migrations form Eurasia.
People have been out of Africa for at least 70,000 years.
During this time some haplogroups formed outside of Africa.
Clyde can you name one to show you are rational?
he origin of a haplogroup is strongly suggested by
a) high frequency
b) high diversity
c) DNA analysis of ancient human remains

quote:The East Indians in Africa, are mainly from North India. They usually believe they are white and during the colonial times they supported Europeans.
Originally posted by the lioness,:
Clyde, were you aware, there are about 3 million Inidan people in Africa, spanning 12 countries

quote:Clyde, stop making up this fake and divisive racial concept of "Black" vs "Mongoloid" native Americans, it's a bad thng you are spreading
Originally posted by Clyde Winters:
Mongoloid Indians in America, have always been a minority because they were usually hunter-gather or nomads. Black Native Americans were farmers and produced abundant surplus food stocks.
quote:I'm puzzled. Why do we have a phylogeny of mtDNA when we are discussing the Y-chromosome?
Originally posted by Troll Patrol # Ish Gebor:
http://www.ianlogan.co.uk/lists/kivisild4.htm
Specificity Origin Lineage(Genesis)
quote:Stupid! How could they continue speaking their Native language when the white man made them slaves or classified them as "free colored" and forced them to learn English.
Originally posted by the lioness,:
quote:Clyde, stop making up this fake and divisive racial concept of "Black" vs "Mongoloid" native Americans, it's a bad thng you are spreading
Originally posted by Clyde Winters:
Mongoloid Indians in America, have always been a minority because they were usually hunter-gather or nomads. Black Native Americans were farmers and produced abundant surplus food stocks.
Even Black people who claim to have Native American ancestry attend the pow wows without contriving these two separate races of native Americans
If the 15 million blacks in this country were not primarily African and instead were primarily Native American many many more would speak Native American language or at least know some words, practice the culture and Europeans would have remarked that Native Americans looked exactly like West Africans, it's a farce you are pulling. Not that some blacks in America don't have Native ancestry but you grossly exaggerte to absurd proportions- and to the detriment of Native Americans.
quote:Are we?
Originally posted by Quetzalcoatl:
quote:I'm puzzled. Why do we have a phylogeny of mtDNA when we are discussing the Y-chromosome?
Originally posted by Troll Patrol # Ish Gebor:
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3322232/bin/gr1.jpg
http://www.ianlogan.co.uk/lists/kivisild4.htm
Specificity Origin Lineage(Genesis)
![[Frown]](frown.gif) If so, then there has been major confusion from the start. I however, tapped in somewhere in the middle of the conversation, where mtDNA Hg M was mentioned.
If so, then there has been major confusion from the start. I however, tapped in somewhere in the middle of the conversation, where mtDNA Hg M was mentioned. quote:--Hong Shi et al. 2008:
The Y chromosome Alu polymorphism (YAP, also called M1) defines the deep-rooted haplogroup D/E of the global Y-chromosome phylogeny [1]. This D/E haplogroup is further branched into three sub-haplogroups DE*, D and E (Figure 1). The distribution of the D/E haplogroup is highly regional, and the three subgroups are geographically restricted to certain areas, therefore informative in tracing human prehistory (Table 1). The sub-haplogroup DE*, presumably the most ancient lineage of the D/E haplogroup was only found in Africans from Nigeria [2], supporting the "Out of Africa" hypothesis about modern human origin. The sub-haplogroup E (E-M40), defined by M40/SRY4064 and M96, was also suggested originated in Africa [3-6], and later dispersed to Middle East and Europe about 20,000 years ago [3,4]. Interestingly, the sub-haplogroup D defined by M174 (D-M174) is East Asian specific with abundant appearance in Tibetan and Japanese (30–40%), but rare in most of other East Asian populations and populations from regions bordering East Asia (Central Asia, North Asia and Middle East) (usually less than 5%) [5-7]. Under D-M174, Japanese belongs to a separate sub-lineage defined by several mutations (e.g. M55, M57 and M64 etc.), which is different from those in Tibetans implicating relatively deep divergence between them [1]. The fragmented distribution of D-M174 in East Asia seems not consistent with the pattern of other East Asian specific lineages, i.e. O3-M122, O1-M119 and O2-M95 under haplogroup O [8,9].
quote:Because you invited people to read your paper on the M haplogroup
Originally posted by Quetzalcoatl:
quote:I'm puzzled. Why do we have a phylogeny of mtDNA when we are discussing the Y-chromosome?
Originally posted by Troll Patrol # Ish Gebor:
http://www.ianlogan.co.uk/lists/kivisild4.htm
Specificity Origin Lineage(Genesis)
quote:
Originally posted by Quetzalcoatl:
For an additional example of Winters's misquoting of sources and refusing to acknowledge that haplotype names change see
https://www.academia.edu/10133899/Clyde_Winters_claim_that_haplotype_M1_proves_that_Indian_Dravidians_are_Mande_speakers_from_Africa_is_not_valid
quote:--Bernard Ortiz de Montellano
In the 1980s and 90s, improvements in the sequencing of DNA led to a number of researchers sequencing the mtDNA of numerous populations in an attempt to trace the evolution of humans and the way in which truly modern humans”, Homo sapiens sapiens, populated Africa and spread out over the globe.
quote:--Catherine Ball
Genetic genealogy is more within reach of the average person than ever, thanks to advances in sequencing technology that have helped the cost of genome sequencing dramatically plummet from nearly $3 billion in 2000 to near $1,000 nowadays. That sort of price reduction is mind-boggling, Ball said. “It’s as if, 15 years from now, I could get my own Mars rover.”
quote:Just as the cost of genetic sequencing has declined the mystery about what DNA can and can not prove has also become evident. In the past we thought that DNA could reliably tell us everything we needed to know about population movements and the history of mankind. As a result, when geneticists claimed this or that we assumed that what they were saying was true and valid.
Originally posted by Troll Patrol # Ish Gebor:
quote:--Bernard Ortiz de Montellano
In the 1980s and 90s, improvements in the sequencing of DNA led to a number of researchers sequencing the mtDNA of numerous populations in an attempt to trace the evolution of humans and the way in which truly modern humans”, Homo sapiens sapiens, populated Africa and spread out over the globe.
quote:--Catherine Ball
Genetic genealogy is more within reach of the average person than ever, thanks to advances in sequencing technology that have helped the cost of genome sequencing dramatically plummet from nearly $3 billion in 2000 to near $1,000 nowadays. That sort of price reduction is mind-boggling, Ball said. “It’s as if, 15 years from now, I could get my own Mars rover.”
http://www.worldsciencefestival.com/2014/05/tracing-family-trees-human-history-genetics/?icn=RA&pos=2
quote:LOL Look at Winters' s impact factor compared to real population geneticists at the bottom of the post. Winter's articles are not cited , by anyone except himself. Clyde please post a citation praising your work in any mainline genetic journal.
Originally posted by Clyde Winters:
[
The goal of lioness and Montellano is to use misdirection to maintain the myth Europeans--whites--carry a unique set of genes that separate them from the African or negro race. It is amazing that my articles on the Dravidians in Current Biology, and my paper on the Fulani in the PNAS, overturned the status quo.
quote:previously posted on
This is why Eurocentrists hate that my articles have been published on-line where everyone can read them. People like lioness and Montellano are useful to establishment researchers in maintaining the status quo. They are useful because real geneticist know that their phylogenies and dating for y-chromosome R and and mtDNA M, are wrong and that these haplogroups had to have been present in Africa before 50kya. As a result, if they attack my work in one of their journals, I would just write a response using their own papers to falsify their research.
That is why Montellano had to publish his paper at Academia edu, instead of a PLoS or PNAS, the genetic evidence proves his article lacks validity.
quote:
Firstly, I am proud to be an Afrocentric scholar. I have been one all my academic life.
LOL. You have to pay a fee to publish in just about all online journals. In most cases as much as $1400 or more .
Moreover, most papers in genetics are published in open access journals.
quote:
You are a joke. I have had articles published in BioEssay, PNAS, Science and etc., these are 'mainstream journals'. Where are your publications?
You can find my population genetics articles here:
http://olmec98.net/archaeogenetics.HTM
quote:Most of the time authors don’t even bother to reply to comments by Winters. When they do, the echo points I’ve made repeatedly on this group.
Subject: RE: editorial policy
Date: Tue, 8 May 2007 07:58:12 -0700
From: "PLoS Genetics" <plosgenetics@plos.org>
To:
Dear Bernard,
Thanks for your message – good question. Reader Responses are intended to be more informal and to encourage community dialogue. As such, they do not undergo peer review by our editors or by external referees (whereas correspondence is treated differently and is peer reviewed).
Instead, Reader Responses are reviewed by staff (to check they are not obscene, abusive, defamatory, libelous, or in some other way illegal or discriminatory; otherwise, we will post them). I hope this helps.
Best wishes,
Andy
Andy Collings
Publications Manager, PLoS Genetics
plosgenetics@plos.org / http://www.plosgenetics.org/
Email Alerts: " target="_blank">http://register.plos.org/
quote:ResearchGate publishes a measure of what impact a particular scholars publications have that includes the number times articles are cited, seen and downloaded together with prestige of the journals they are published in equaling an impact factor
MtDNA-based genetic arguments provided by Dr. Winters in favor of gene flow from Africa to Dravidian-speaking Indians are, however, entirely erroneous. The author has been, unfortunately, confused by overlooking changes in mtDNA haplogroup (hg) nomenclature. Namely hg, M1 in Kivisild et al.(4) has been later changed to hg M3, in order to avoid parallel nomenclatures.(5) Furthermore, a recent dedicated paper on phylogeography of mtDNA hg M1(6) as well as an extensive comparative mapping of autosomal genetic markers among many Indian populations relative to global populations elsewhere, including Africans,(7) do not provide any clues for a putative recent gene flow, from Africa, to Dravidian-speaking populations in South Asia.
quote:This is my response to Oppenheimer
Originally posted by Quetzalcoatl:
A post-Review of a "paper" by Clyde Winters
http://classic.rstb.royalsocietypublishing.org/content/367/1590/770.abstract/reply - content-block
Haplogroup L3 (M,N) probably spread across Africa before the Out of Africa event
• Clyde Winters, Professor
Uthman dan Fodio Institute
Oppenheimer presents a convincing article supporting an Out of Africa (OoA) event [1]. At this time, beachcombing anatomically modern humans (amh) exploiting marine resources made their way to Australia by 42kya, after a bottleneck in India [1]. It was in Eurasia that, according to Oppenheimer, L3 (M,N) originated and haplogroups M and N returned to Africa via a back migration [1]. Soares et al. also argue that L3 (M,N and R) originated in Eurasia [2].
Oppenheimer dates L3 (M,N) to 83 kya [1], while Soares et al. [2] and Behar [3] argue that L3 originated around 70 kya and haplogroups M,N, and R appeared between 50-59 kya in Eurasia.
The genetic phylogeny of L3 suggest that L3 (M,N) had probably spread across Africa before the Toba eruption and OoA event so it could not have originated in Eurasia. Haplogroup L3 (M,N) probably spread from East Africa to West Africa along with haplogroup LOd.
The most recent common ancestor (TMRC) of AMH carrying LOd according to Gonder et al. dates to 106kya. A haplotype of LOd is AF-24, delineated by DdeI site 10394 and AluI site of np 10397. Haplogroup L3 (M,N) is characterized by the DdeI site np 10394 and AluI 10397. Chen et al. maintain that Haplotype AF-24 (DQ112852) is at the base of the M Haplogroup [4].
The only African mtDNA that has both of these sites is the Senegalese AF-24 associated with Haplogroup L3 [Chen]. The presence of AF-24 among the Senegalese, given the antiquity of this haplotype in relation to LOd, suggests an early expansion of L3 (M,N) from East Africa to West Africa before the OoA event suggested by Oppenheimer.
Gonder et al. has dated L3 to 100kya [5]. Oppenheimer [1] and Behar et al [3] on the other hand have postulated an estimated time for TMRCA of L3 to around 70kya. The presence of L3 (M,N) in West Africa and haplotype AF- 24 suggest an ancient demic diffusion of L3 (M,N) to West Africa prior to 70kya, and support Soares et al.'s [2] and Gonder et al.'s [5] dating of L3 between 80-100kya.
Anatomically modern humans arrived in Senegal during the Sangoan period. Sangoan artifacts spread from East Africa to West Africa between 100-80kya. In Senegal Sangoan material was discovered near Cap Manuel [6], Gambia River in Senegal [8,9]; and Cap Vert [7]. The distribution of the Sangoan culture supports the demic diffusion of L3 (M,N) into West Africa over 100kya.
The spread of the Sangoan tool kit into the Senegambian region [6-9] would explain the presence of haplotype AF-24 among the Senegalese. This archaeological evidence adds additional corroboration to the West African genetic phylogenies which point to the probable expansion of L3(M,N) from East to West Africa prior to the OoA event indicated by Oppenheimer [1] 100 kya.
References:
1. Oppenheimer S. 2012 Out-of-Africa, the peopling of continents and islands: tracing uniparental gene trees across the map. Phil. Trans. R. Soc. Lond. B, 367, 770-784. (doi: 10.1098/rstb.2011.0306)
2. Soares P., Ermini L., Thomson, N., Mormina M., Rito T., Rohl A., Salas A., Oppenheimer S., Macaulay V., Richards M.B. 2009 Correcting for purifying selection: an improved human mitochondrial molecular clock. Am. J. Hum. Genet., 84, 740-759. (doi:10.1016/j.ajhg.2009.05.001)
3. Oppenheimer S. 2009 The great arc of dispersal of modern humans: Africa to Australia. Quat. Int., 202, 2-13. (doi:10.1016/j.quaint.2008.05.015)
4. Chen Y-S., Olckers A., Schurr T.G., Kogelnik A.M., Huoponen K., Wallace D.C. 2000 mtDNA variation in the South African Kung and Khwe - and their genetic relationships to other African populations. Am. J. Hum. Genet., 66, 1362-1383.
5. Gonder M.K., Mortensen H.M., Reed F.A., de Sousa A., Tishkoff S.A. 2007 Whole mtDNA genome sequence analysis of ancient African lineages. Mol. Biol. Evol., 24, 757-768. (doi: 10.1093/molbev/msl209)
6. Giresse, P. 2008 Tropical and sub-Tropical West Africa - marine and continental changes during the Late Quaternary. Elsevier Science.
7. Phillipson, D.W. 2005 African Archaeology. Cambridge University Press, Cambridge.
8. Davies, O. 1967 West Africa before the Europeans. Methuen, London.
9. Wai-Ogusu, A. 1973 Was there a Sangoan industry in West Africa. West Afr. J. Arcah., 3, 191-96.
%%%%%%%%%%%%
Response to Winters (2012) 'Haplogroup L3 (M,N) probably spread across Africa before the Out of Africa event'
• Stephen Oppenheimer
School of Anthropology and Museum Ethnography, Institute of Cognitive and Evolutionary Anthropology,
Winters' response to my recent review in Phil. Trans. R. Soc. B [1] argues that the two mtDNA super-haplogroups M and N (which account for nearly all non-Africans and derive from African haplogroup L3) both originated within Africa, before the anatomically modern human (AMH) 'Out of Africa' (OOA) exit, and further that African L3 dates to 100kya (the latter date being based on a misreading of the cited sources). His argument rests mainly on the acknowledged phylogenetic ambiguity of a single Senegalese sample in a paper published in 2000 before complete sequencing was routinely used to resolve such ambiguities.
Local African origins of both N & M have been argued before on the basis of the presence of M sub-group M1 and certain derivative N sub- groups in East Africa; and this evidence of local backflow has already been clarified in the modern literature - as discussed in my review [1]. While the said N sub-groups are clearly derived from N outside Africa and are much younger than any postulated exit date, African sub-group M1 is, perhaps, more interesting having a rather greater age than N in Africa, though still considerably younger than any estimate of the OOA event [1] and M1 is present, though rare [2], outside Africa, consistent with an ex- African origin.
Genetic phylogeographic arguments contribute greatly to the topographical inference of the sequence of migrations and, independently of any date estimates, argue strongly for M & N originating outside Africa from a single L3 exit [1]. MtDNA dates are presently more reliable than for any other genetic locus but, as discussed exhaustively in my review [1], their high calculated variance still gives wide error bars and the opportunity for much fruitless argument on precise dates of AMH exit either side of the Toba volcanic event, which could best be resolved by diagnostic, accurately dated modern human fossils (rather than stone tools) at key points outside Africa.
For the dates suggested by Winters, such early AMH fossils are currently lacking. As for the chosen genetic dates he cites for his own preferred chronology, several of which are misquoted and/or misunderstood, those from the older literature have been superseded by more recent, more resolved, recalibrated, comprehensive, accurate and precise studies on L3 within and outside Africa, including M and N [3, 4], including one major source Winters suggests himself [5].
Finally, there are is a string of misunderstandings mistakes and misquotes in Winters' letter listed below, which detract from, rather than supporting, his overall argument:
Para 1, sentence 2:
42 kya refers presumably to the earliest carbon dates in Timor, not those much earlier luminescence dates given, in my review [1], for Australia.
Para 2, sentence 1: 'Oppenheimer dates L3 (M,N) to 83 kya'
Comment: I did not do so in this review [1]. As explained in the text I chose throughout to cite lineage ages from the key mtDNA recalibration paper of Soares et al. (2009) [5]. This was both for consistency, and to use the latest, most comprehensive and, hopefully, least inaccurate method, rather than pick and choose results from older, phylogenetically less-resolved publications, which might perhaps have suited my own preference for a pre-Toba exit better.
Para 3 (whole para): Comment: No published evidence/reference given for these assertions.
Para 4, sentence 1: 'The most recent common ancestor (TMRC) of AMH carrying LOd according to Gonder et al. dates to 106kya.'.
Comment: 'LOd' is not 'TMRC of AMH' (nor is L0d).
Para 4, sentence 1: 'A haplotype of LOd is AF-24...'
Comment: No it is not - on the evidence given in the citation. Gonder et al. (2007) [6] do not even mention haplotype AF-24 as such. The claim, if it were true, would simply reinforce the impression of ambiguity. AF-24 is however mentioned by Winters' other cited reference (published in 2000 and not based on complete sequences): Chen et al. (2000) [7] shown as belonging to L3a; but those authors acknowledge this particular phylogenetic assignment to be poorly resolved:
'Haplotype AF24, which is aligned with Asian macrohaplogroup M, is indicated by a double section symbol...subclusters AF19-AF21/AF24 and AF80-AF84 were not resolved at bootstrap values >50%...' [7]
They further emphasize the ambiguity of its phylogenetic/geographic assignment:
'...it is also possible that this particular haplotype [AF-24] is present in Africa because of back-migration [of M] from Asia.'... and: 'Alternatively, AF24 may have been introduced from Asia into Africa more recently.' [7]
Para 4, sentence 3: Winters continues to mis-cite Chen et al. (2000) [7]:
'Chen et al. maintain that Haplotype AF-24 (DQ112852) is at the base of the M Haplogroup [4].', Comment: Where? - unless it is in their reference to back-migration (above).
Para 5 appears to be further argument based on phylogenetic ambiguity.
Para 6, sentence 1: 'Gonder et al. has dated L3 to 100kya (5).'
Comment: No, they do not, according to Gonder's Table 2 [6].
Para 6, sentence 3: 'The presence of L3 (M,N) in West Africa and haplotype AF- 24 suggest an ancient demic diffusion of L3 (M,N) to West Africa prior to 70kya, and support Soares et al.'s (2) and Gonder et al.'s (5) dating of L3 between 80-100kya.'
Comment: The above inference, based on a single poorly-resolved haplotype, is unsound and Soares et al. (2009) [5] are mis-cited as far as the date is concerned.
Paras 7 and 8: Varied, inadequately-cited references to the presence of the 'Sangoan tool kit' in West Africa are used by Winters to infer the movement and spread of 'L3 (M,N)' in West Africa.
Comment: This is an unwarranted inference using as it does, hypothetical links (for which no evidence is given) between an Early Stone Age cultural phase and an ambiguous single modern genetic haplotype (AF-24). While dated archaeological evidence of human presence can, occasionally, be used to infer first-ever human arrival in a previously uninhabited region e.g. Polynesia or the Canary Islands and this kind of evidence be used to cross -check calibration of the mtDNA clock on unique and specific local founding lineages in those places [5], the sort of "stones and genes" type of inferences Winters makes for the spread of 'L3 (M,N)' in West Africa are completely unwarranted.
References
1. Oppenheimer S. 2012 Out-of-Africa, the peopling of continents and islands: tracing uniparental gene trees across the map. Phil. Trans. R. Soc. Lond. B, 367, 770-784. (doi: 10.1098/rstb.2011.0306)
2. Gunnarsdottir E.D., Nandineni M.R., Li M., Myles S., Gil D., Pakendorf B., Stoneking M. (2011) Larger mitochondrial DNA than Y- chromosome differences between matrilocal and patrilocal groups from Sumatra. Nature Communications, 2, 228. (doi: 10.1038/ncomms1235)
3. Soares P., Alshamali F., Pereira J.B., Fernandes, V., N.M. Silva, Afonso C., Costa M.D., Musilova E., Macaulay V., Richards M.B., Cerny V., Pereira Luisa. (2012) The expansion of mtDNA haplogroup L3 within and out of Africa. Mol. Biol. Evol., 29, 915-927. (doi: 10.1093/molbev/msr245)
4. Fernandes, V., Alshamali F., Alves M., Costa M.D., Pereira J.B., N.M. Silva, Cherni L., Harich N., Cerny V., Soares P. Richards M.B., Pereira Luisa. (2012) The Arabian cradle: Mitochondrial relicts of the first steps along the Southern Route out of Africa. Am. J. Hum. Genet., 90, 347-355. (doi: 10.1016/j.ajhg.2011.12.010)
5. Soares P., Ermini L., Thomson, N., Mormina M., Rito T., Rohl A., Salas A., Oppenheimer S., Macaulay V., Richards M.B. 2009 Correcting for purifying selection: an improved human mitochondrial molecular clock. Am. J. Hum. Genet., 84, 740-759. (doi:10.1016/j.ajhg.2009.05.001)
6. Gonder M.K., Mortensen H.M., Reed F.A., de Sousa A., Tishkoff S.A. 2007 Whole mtDNA genome sequence analysis of ancient African lineages. Mol. Biol. Evol., 24, 757-768. (doi: 10.1093/molbev/msl209)
7. Chen Y-S., Olckers A., Schurr T.G., Kogelnik A.M., Huoponen K., Wallace D.C. 2000 mtDNA variation in the South African Kung and Khwe - and their genetic relationships to other African populations. Am. J. Hum. Genet., 66, 1362-1383.
quote:You're just jealous that I have peer reviewed articles published eventhough I am an Afrocentric researcher.It is great news that Professional Scientists in India and Pakistan have the nerve to publish their own peer reviewed journals.
Originally posted by Quetzalcoatl:
quote:LOL Look at Winters' s impact factor compared to real population geneticists at the bottom of the post. Winter's articles are not cited , by anyone except himself. Clyde please post a citation praising your work in any mainline genetic journal.
Originally posted by Clyde Winters:
[
The goal of lioness and Montellano is to use misdirection to maintain the myth Europeans--whites--carry a unique set of genes that separate them from the African or negro race. It is amazing that my articles on the Dravidians in Current Biology, and my paper on the Fulani in the PNAS, overturned the status quo.
quote:previously posted on
This is why Eurocentrists hate that my articles have been published on-line where everyone can read them. People like lioness and Montellano are useful to establishment researchers in maintaining the status quo. They are useful because real geneticist know that their phylogenies and dating for y-chromosome R and and mtDNA M, are wrong and that these haplogroups had to have been present in Africa before 50kya. As a result, if they attack my work in one of their journals, I would just write a response using their own papers to falsify their research.
That is why Montellano had to publish his paper at Academia edu, instead of a PLoS or PNAS, the genetic evidence proves his article lacks validity.
http://www.egyptsearch.com/forums/ultimatebb.cgi?ubb=get_topic;f=15;t=005796;p=1
Most of the papers Winters is posting have been published in journals from India or Pakistan which are post-reviewed i.e. no one reviews them before publication (no, they don't check the references either). People reading the paper are supposed to write and critique it afterwards LOL. It's not worth the effort. Winter has yet to publish a peer-reviewed publication in a recognized mainline scientific journal. BTW letters to the editor and comments to other published articles are NOT peer-reviewed. People, who have published in peer-reviewed journals and are familiar with the process know the difference.
Winters:
quote:
Firstly, I am proud to be an Afrocentric scholar. I have been one all my academic life.
LOL. You have to pay a fee to publish in just about all online journals. In most cases as much as $1400 or more .
Moreover, most papers in genetics are published in open access journals.
Not true, most of the influential and cited articles in genetics are published in mainline peer-reviewed journals for instance just a few of the ones I look at American Journal of Human Genetics, Annals of Human Genetics[,[Proceedings of the National Academy of Science, PNAS, European Journal of Genetics, Nature, Science, Nature Genetics,Nature Reviews-Genetics;[BMC Genetics ,BMC Genomics, Molecular Biology and Evolution, American Journal of Physical Evolution, etc. etc. this is just a tiny sample of the peer-reviewed journals on genetics- take a look at the list in "Entrez PubMed."
Winters:
quote:
You are a joke. I have had articles published in BioEssay, PNAS, Science and etc., these are 'mainstream journals'. Where are your publications?
You can find my population genetics articles here:
http://olmec98.net/archaeogenetics.HTM
I have published in a couple of papers in Science and Current Anthropology as well as papers in Journal of the American Anthropological Society. Journal of the American Chemical Society, Ethnohistory, Medical Anthropology,Advances in Nursing Science,Yearbook of Physical Anthropology,Journal of Ethnomedicine, Ethnomedizin among others as well as peer reviewing dozens of papers for mainline peer-reviewed journals.
Analyzing Winters’ list of publications confirms my previous statement that he has not published a refereed article in a mainline refereed journal. There were a number of broken links in his list, but all of his contributions to mainline journals i.e. Bioassay, PNAS, Science, and PLoS Genetics were letters commenting on someone else’s article. Letters and comments are NOT peer-reviewed, and are mostly ignored by the author’s of the original papers. Winters is probably counting on the fact that most people are not familiar with academic publishing and can be bamboozled into thinking that his contributions to prestigious journals are peer-reviewed articles. Don’t take my word, go to his list and verify that these contributions are letters.
I wrote the editors of the mainline journals where Winters published his comments, and as expected these things are NOT peer-reviewed before publication. For example from PLoS Genetics
quote:Most of the time authors don’t even bother to reply to comments by Winters. When they do, the echo points I’ve made repeatedly on this group.
Subject: RE: editorial policy
Date: Tue, 8 May 2007 07:58:12 -0700
From: "PLoS Genetics" <plosgenetics@plos.org>
To:
Dear Bernard,
Thanks for your message – good question. Reader Responses are intended to be more informal and to encourage community dialogue. As such, they do not undergo peer review by our editors or by external referees (whereas correspondence is treated differently and is peer reviewed).
Instead, Reader Responses are reviewed by staff (to check they are not obscene, abusive, defamatory, libelous, or in some other way illegal or discriminatory; otherwise, we will post them). I hope this helps.
Best wishes,
Andy
Andy Collings
Publications Manager, PLoS Genetics
plosgenetics@plos.org / http://www.plosgenetics.org/
Email Alerts: " target="_blank">http://register.plos.org/
“Did the Dravidian Speakers Originate in Africa” In a familiar pattern, the authors of the article Winters was commenting on point out that Winters is not familiar with genetics:
Chaubey, G. et al. 2007 “Reply to Winters,” BioAssays 29: 499.quote:ResearchGate publishes a measure of what impact a particular scholars publications have that includes the number times articles are cited, seen and downloaded together with prestige of the journals they are published in equaling an impact factor
MtDNA-based genetic arguments provided by Dr. Winters in favor of gene flow from Africa to Dravidian-speaking Indians are, however, entirely erroneous. The author has been, unfortunately, confused by overlooking changes in mtDNA haplogroup (hg) nomenclature. Namely hg, M1 in Kivisild et al.(4) has been later changed to hg M3, in order to avoid parallel nomenclatures.(5) Furthermore, a recent dedicated paper on phylogeography of mtDNA hg M1(6) as well as an extensive comparative mapping of autosomal genetic markers among many Indian populations relative to global populations elsewhere, including Africans,(7) do not provide any clues for a putative recent gene flow, from Africa, to Dravidian-speaking populations in South Asia.
here are the scores for the four authors of the reply above, the author of the review of Winters’s article starting this thread as well as Winter’s and my impact factor for comparison:
Name papers reads citations impact
Toomas Kivisild 157 15K 9,444 1,133.89
R. Villems 179 15K 8,485 1,122.55
Mait Metzpalu 69 12K 2,870 665.13
G. Chaubey 53 6K 1,020 322.03
Klyosov, A. 96 3K 1,168 141.94
Bernard O de M 45 521 348 223.18
Clyde Winters 65 3K 92 56.22
quote:Yes, from my paper in Academia.edu
Originally posted by Troll Patrol # Ish Gebor:
@ Quetzalcoatl,
Are these the papers you are referring at, when you spoke of the M1-M3 transposition?
4. Kivisild T, Kaldma K, Metspalu M, Parik J, Papiha S, Villems R. 1999. The place of the Indian mtDNA variants in the global network of maternal lineages and the peopling of the Old World. In Deka R, Papiha SS. eds Genomic Diversity Kluwer Academic/Plenum Publishers New York p 135–152.
5. Quintana-Murci L, Semino O, Bandelt H-J., Passarino G, McElreavey K, Santachiara-Benerecetti AS. 1999. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet 23: 437 – 441.
quote:
. Kivisild (1999) and Quintana-Murci (1999) identified and named different haplotypes, one Indian and one African, as M1. This happens often and the usual procedure is for one of the authors to change the nomenclature. The final arbiter is www.phylotree.org that publishes an updated mtDNA tree. In this case, Quintana-Murci’s (1999) identification of M1 was retained with haplotype markers 16129 16189 16249 16311. Kivisild’s M1 had 16126 as the marker and this haplogroup’s name was changed to M3. The individuals in Kivisild (1999) labeled M1 did not disappear their designation changed to M3. Kivisild’s compliance with the revised nomenclature is seen in Bamshad (2001). Numerous papers including some cited by Winters make it clear that there is no M1 in India although M3 with the 16126 marker (1999’s M1) is definitely found (Bamshad 2001; Maca-Meyer 2001; Kivisild 2003- Dravidians; Metspalu 2004; Rajkumar 2005; Thangaraj 2006) Chandrasekar 2009; Shah 2012). In 2007, Winters (2007) repeated this distortion in a comment to a paper by Chaubey (2006). Chaubey, Metspalu, Kivisild, and Villems (2007) replied and clearly pointed out Winters’ misreporting of their paper. In 2008, Kivisild and Villems pointed this error to Winters by e-mail. Yet, he continues to use this argument and citation. This is academic dishonesty.
References
Bamshad, M, T. Kivisild, et al. 2001 "Genetic Evidence on the Origins
of Indian Caste Populations," Genome Research 11: 994-1004
Chaubey, G., Metspalu, M., Villems, M. R., Kivisild, T. 2007. “Reply to Winters” BioEssays 29(5): 499 Published Online: 20 Apr 2007
DOI: 10.1002/bies.20574.
Chaubey,G, Metspalu, M., Kivisild, T. and Villems, R. 2006 ”Peopling of South Asia: Investigating the Caste-Tribe Continuum in India,” Bio
Essays 29:91-100.
Chandrasekar, et al. 2009 “Updating Phylogeny of Mitochondrial DNA Macrohaplogroup M in India: Dispersal of Modern Human in South Asian Corridor,” PLoS ONE 4(10)e7447.
Kivisild, T. et al. 1999a “Deep common ancestry of Indian and western-Eurasian mitochondrial DNA lineages, Current Biology 9:1331–1334.
Kivisild, T. et al. 1999b “The Place of the Indian mtDNA Variants in the Global Network of Maternal Lineages and the Peopling of the Old World,” In Deka, R.P. ed. Genomic Diversity, 135-152 NY: S.S. Kluwer/Plenum Publishers.
Kivisild, T., et al. 2003 “The Genetic Heritage of the Earliest Settlers Persists Both in Indian Tribal and Caste Populations,” American Journal of Human Genetics 72(2): 313-332
Maca-Meyer, N., et al. 2001 “Major Genomic Mitochondrial Lineages Delineate Early Human Expansions,” BMC Genetics 2:13.
Metspalu, M., et al. 2004 “More of the Extant mtDNA Boundaries in South and Southwest Asia were Likely Shaped During the Initial
Settlement of Eurasia by Anatomically Modern Humans,” BMC Genetics 5:26
Quintana-Murci, L., et al. 1999 "Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa," Nature Genetics 23: 437-441.
Rajkumar, R., et al. 2005 “Phylogeny and Antiquity of M Macrohaplogroup Inferred from Complete mtDNA Sequence of Indian Specific Lineages,” BMC Evolutionary Biology 5:26.
Sharma, G., et al. 2012 “Genetic Affinities of the Central Indian Tribal Populations,” PLoS ONE 7(2):e35246.
quote:Science is universal and does not have a color contingent. Scientists from all over the world (with all hues of skin color) publish in the mainline refereed journals. Here is a list of Nobel prize winners by country of birth
Originally posted by Troll Patrol # Ish Gebor:
Quetzalcoatl, is like. It's only peer reviewed if (whites) the western world has reviewed and consigned it.
It's somewhat the mindset of a colonist.
What Quetzalcoatl is saying is that western academia is superior, therefore credible.
quote:I agree on that, but why did you judge those scientists from India and Pakisan?
Originally posted by Quetzalcoatl:
quote:Science is universal and does not have a color contingent. Scientists from all over the world (with all hues of skin color) publish in the mainline refereed journals. Here is a list of Nobel prize winners by country of birth
Originally posted by Troll Patrol # Ish Gebor:
Quetzalcoatl, is like. It's only peer reviewed if (whites) the western world has reviewed and consigned it.
It's somewhat the mindset of a colonist.
What Quetzalcoatl is saying is that western academia is superior, therefore credible.
quote:So, M1 became M3 somewhere in 1999.
Originally posted by Quetzalcoatl:
quote:Yes, from my paper in Academia.edu
Originally posted by Troll Patrol # Ish Gebor:
@ Quetzalcoatl,
Are these the papers you are referring at, when you spoke of the M1-M3 transposition?
4. Kivisild T, Kaldma K, Metspalu M, Parik J, Papiha S, Villems R. 1999. The place of the Indian mtDNA variants in the global network of maternal lineages and the peopling of the Old World. In Deka R, Papiha SS. eds Genomic Diversity Kluwer Academic/Plenum Publishers New York p 135–152.
5. Quintana-Murci L, Semino O, Bandelt H-J., Passarino G, McElreavey K, Santachiara-Benerecetti AS. 1999. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet 23: 437 – 441.
quote:
. Kivisild (1999) and Quintana-Murci (1999) identified and named different haplotypes, one Indian and one African, as M1. This happens often and the usual procedure is for one of the authors to change the nomenclature. The final arbiter is www.phylotree.org that publishes an updated mtDNA tree. In this case, Quintana-Murci’s (1999) identification of M1 was retained with haplotype markers 16129 16189 16249 16311. Kivisild’s M1 had 16126 as the marker and this haplogroup’s name was changed to M3. The individuals in Kivisild (1999) labeled M1 did not disappear their designation changed to M3. Kivisild’s compliance with the revised nomenclature is seen in Bamshad (2001). Numerous papers including some cited by Winters make it clear that there is no M1 in India although M3 with the 16126 marker (1999’s M1) is definitely found (Bamshad 2001; Maca-Meyer 2001; Kivisild 2003- Dravidians; Metspalu 2004; Rajkumar 2005; Thangaraj 2006) Chandrasekar 2009; Shah 2012). In 2007, Winters (2007) repeated this distortion in a comment to a paper by Chaubey (2006). Chaubey, Metspalu, Kivisild, and Villems (2007) replied and clearly pointed out Winters’ misreporting of their paper. In 2008, Kivisild and Villems pointed this error to Winters by e-mail. Yet, he continues to use this argument and citation. This is academic dishonesty.
References
Bamshad, M, T. Kivisild, et al. 2001 "Genetic Evidence on the Origins
of Indian Caste Populations," Genome Research 11: 994-1004
Chaubey, G., Metspalu, M., Villems, M. R., Kivisild, T. 2007. “Reply to Winters” BioEssays 29(5): 499 Published Online: 20 Apr 2007
DOI: 10.1002/bies.20574.
Chaubey,G, Metspalu, M., Kivisild, T. and Villems, R. 2006 ”Peopling of South Asia: Investigating the Caste-Tribe Continuum in India,” Bio
Essays 29:91-100.
Chandrasekar, et al. 2009 “Updating Phylogeny of Mitochondrial DNA Macrohaplogroup M in India: Dispersal of Modern Human in South Asian Corridor,” PLoS ONE 4(10)e7447.
Kivisild, T. et al. 1999a “Deep common ancestry of Indian and western-Eurasian mitochondrial DNA lineages, Current Biology 9:1331–1334.
Kivisild, T. et al. 1999b “The Place of the Indian mtDNA Variants in the Global Network of Maternal Lineages and the Peopling of the Old World,” In Deka, R.P. ed. Genomic Diversity, 135-152 NY: S.S. Kluwer/Plenum Publishers.
Kivisild, T., et al. 2003 “The Genetic Heritage of the Earliest Settlers Persists Both in Indian Tribal and Caste Populations,” American Journal of Human Genetics 72(2): 313-332
Maca-Meyer, N., et al. 2001 “Major Genomic Mitochondrial Lineages Delineate Early Human Expansions,” BMC Genetics 2:13.
Metspalu, M., et al. 2004 “More of the Extant mtDNA Boundaries in South and Southwest Asia were Likely Shaped During the Initial
Settlement of Eurasia by Anatomically Modern Humans,” BMC Genetics 5:26
Quintana-Murci, L., et al. 1999 "Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa," Nature Genetics 23: 437-441.
Rajkumar, R., et al. 2005 “Phylogeny and Antiquity of M Macrohaplogroup Inferred from Complete mtDNA Sequence of Indian Specific Lineages,” BMC Evolutionary Biology 5:26.
Sharma, G., et al. 2012 “Genetic Affinities of the Central Indian Tribal Populations,” PLoS ONE 7(2):e35246.
quote:I just copied what the web site had. They seem to list countries that had changed names. For example Belarus at one time was an SSR in the Soviet conglomerate. Is this what you were curious about?
Originally posted by Troll Patrol # Ish Gebor:
quote:I agree on that, but why did you judge those scientists from India and Pakisan?
Originally posted by Quetzalcoatl:
quote:Science is universal and does not have a color contingent. Scientists from all over the world (with all hues of skin color) publish in the mainline refereed journals. Here is a list of Nobel prize winners by country of birth
Originally posted by Troll Patrol # Ish Gebor:
Quetzalcoatl, is like. It's only peer reviewed if (whites) the western world has reviewed and consigned it.
It's somewhat the mindset of a colonist.
What Quetzalcoatl is saying is that western academia is superior, therefore credible.
quote:In 1999 potential confusion arose because 2 papers were published that called certain sequences M1. One had to change names, and Phylotree designated that the one carrying M16126 would now be named M3.
Originally posted by Troll Patrol # Ish Gebor:
quote:So, M1 became M3 somewhere in 1999.
Originally posted by Quetzalcoatl:
quote:Yes,
Originally posted by Troll Patrol # Ish Gebor:
[qb] @ Quetzalcoatl,
Are these the papers you are referring at, when you spoke of the M1-M3 transposition?
4. Kivisild T, Kaldma K, Metspalu M, Parik J, Papiha S, Villems R. 1999. The place of the Indian mtDNA variants in the global network of maternal lineages and the peopling of the Old World. In Deka R, Papiha SS. eds Genomic Diversity Kluwer Academic/Plenum Publishers New York p 135–152.
5. Quintana-Murci L, Semino O, Bandelt H-J., Passarino G, McElreavey K, Santachiara-Benerecetti AS. 1999. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet 23: 437 – 441.
The question I'm having here is, what is the "new" M = M1 reference?
quote:As you can see I did admit that only 5.2% of the pygmy carried R1.
Y-chromosome R1 is found throghout Africa. The pristine
form of R1-M173 is only found in Africa (Coia et al,
2005; Cruciani et al, 2002, 2010). The age of ychromosome
R is 27ky. Most researchers believe that
R(M173) is 18.5 ky.There is a great diversity of the
macrohaplogroup R in Africa (See Figure 1). Ychromosome
R is characterized by M207/V45. The V45
mutation is found among African populations ( Cruciani et
al ,2010). ISOGG 2010 Y-DNA haplogroup tree makes it
clear that V45 is phylogenetically equivalent to M207.The
most common R haplogroup in Africa is R1 (M173). The
predominant haplogroup is R1b (Berniell-Lee et al,2009;
Coia et al, 2005; Winters, 2010b; Wood et al, 2009).
Cruciani et al (2010) discovered new R1b mutations
including V7, V8, V45, V69, and V88. Geography appears
to play a significant role in the distribution of haplogroup
R in Africa. Cruciani et al (2010) has renamed the R*-
M173 (R P-25) in Africa V88. The TMRCA of V88 was
9200-5600 kya (Cruciani et al, 2010).
Y-chromosome V88 (R1b1a) has its highest frequency
among Chadic speakers, while the carriers of V88 among
Niger-Congo speakers (predominately Bantu people)
range between 2-66% ( Cruciani et al, 2010; Bernielle-Lee
et al, 2009). Haplogroup V88 includes the mutations M18,
V35 and V7. Cruciani et al (2010) revealed that R-V88 is
also carried by Eurasians including the distinctive
mutations M18, V35 and V7.
R1b1-P25 is found in Western Eurasia. Haplogroup
R1b1* is found in Africa at various frequencies. BerniellLee
et al (2009) found in their study that 5.2% carried
Rb1*. The frequency of R1b1* among the Bantu ranged
from 2-20. The bearers of R1b1* among the Pygmy
populations ranged from 1-25% (Berniell-Lee et al, 2009).
The frequency of R1b1 among Guinea-Bissau populations
was 12% (Carvalho et al,2010).
quote:As a result, I did not attempt to decieve anyone about the frequency of R1 in Africa as the author implies.
Around 0.1 of Sub Saharan Africans carry R1b1b2. Wood
et al (2009) found that Khoisan (2.2%) and Niger-Congo
(0.4%) speakers carried the R-M269 y-chromosome.
The Khoisan also carry RM343 (R1b) and M 198 (R1a1)
(Naidoo et al., 2010) the archaeological and linguistic data
indicate the successful colonization of Asia by SubSaharan
Africans from Nubia 5-4kya (Winters, 2007,2008,
2010c). The archaeological evidence makes it clear that
around 4kya intercultural style artifacts connected Africa
and Eurasia (Winters, 2007,2010c).




quote:This argument is invalid. It implies that Phylotree promotes lies to maintain the status quo, and white supremacy.
Originally posted by Quetzalcoatl:
In 1999 potential confusion arose because 2 papers were published that called certain sequences M1. One had to change names, and Phylotree designated that the one carrying M16126 would now be named M3.
M1 16129 16189 16249 16311
M3 16126
Thus, it is absolutely clear that, at least since the year 2000, mutation at 16126 has designated haplogroup M3 and mutation at 16129 designates haplogroup M1.
This is what you see if you visit phylotre- the official naming site.
Is this what you questioned?

quote:Are these Researchers who maintain this white?
Originally posted by Clyde Winters:
Clearly Phylotree is unreliable and based on Euroocentrism and white supremacy. This is obvious given the way R-M173 in Africa was changed into V88, and 207 in Africa was changed into V45. All of these efforts are simply misdirection aimed at containing Negroes in Africa, and separating Dravidian speakers from Africans.
Researchers maintain that the Eastern African HVS-I signature motifs are 16,129, 16,189, 16,249, and 16,311. This motiff is found in Indian M1, so it can not be changed into M3, because M3 already existed.
.
quote:This is pathetic. AS usual, when the facts and the data have you trapped, you immediately resort to ad hominem and conspiracy theories. Thousands of scientists around the word do their research on many many areas and could care less than a fig about, you, Afrocentrism, or Hindutva. Every Science has an organization that is in charge of regulating nomenclature and preventing mixups ; for example, in my own field the International Union of Pure and Applied Chemistry (IUPAC) sets the rules for naming chemical compounds. And Phylotree is the official site for naming and organizing Y-chromosome and mtDNA trees.
Originally posted by Clyde Winters:
quote:This argument is invalid. It implies that Phylotree promotes lies to maintain the status quo, and white supremacy.
Originally posted by Quetzalcoatl:
In 1999 potential confusion arose because 2 papers were published that called certain sequences M1. One had to change names, and Phylotree designated that the one carrying M16126 would now be named M3.
M1 16129 16189 16249 16311
M3 16126
Thus, it is absolutely clear that, at least since the year 2000, mutation at 16126 has designated haplogroup M3 and mutation at 16129 designates haplogroup M1.
This is what you see if you visit phylotre- the official naming site.
Is this what you questioned?
Clearly Phylotree is unreliable and based on Euroocentrism and white supremacy. This is obvious given the way R-M173 in Africa was changed into V88, and 207 in Africa was changed into V45. All of these efforts are simply misdirection aimed at containing Negroes in Africa, and separating Dravidian speakers from Africans.
Researchers maintain that the Eastern African HVS-I signature motifs are 16,129, 16,189, 16,249, and 16,311. This motiff is found in Indian M1, so it can not be changed into M3, because M3 already existed.
.
.
The presence of one transition, 126, in Indian M1, does not deny its existence as M1, since it is the only M haplogroup outside of Africa that carries the entire M1 motiff. Luckily, we have the original paper that proves that M1 in India exist.
.
quote:Yes you stated the facts clearly. Someone at Phylotree decided to name M1 in India, M3. Just because someone changed the name for Indian M1, does not erase M1 from India. You admit yourself that Phylotree is in charge of regulating nomenclature . Phylotree is not trying to prevent mixups, they are just trying to maintain the status quo and white supremacy.
Originally posted by Quetzalcoatl:
quote:This is pathetic. AS usual, when the facts and the data have you trapped, you immediately resort to ad hominem and conspiracy theories. Thousands of scientists around the word do their research on many many areas and could care less than a fig about, you, Afrocentrism, or Hindutva. Every Science has an organization that is in charge of regulating nomenclature and preventing mixups ; for example, in my own field the International Union of Pure and Applied Chemistry (IUPAC) sets the rules for naming chemical compounds. And Phylotree is the official site for naming and organizing Y-chromosome and mtDNA trees.
Originally posted by Clyde Winters:
quote:This argument is invalid. It implies that Phylotree promotes lies to maintain the status quo, and white supremacy.
Originally posted by Quetzalcoatl:
In 1999 potential confusion arose because 2 papers were published that called certain sequences M1. One had to change names, and Phylotree designated that the one carrying M16126 would now be named M3.
M1 16129 16189 16249 16311
M3 16126
Thus, it is absolutely clear that, at least since the year 2000, mutation at 16126 has designated haplogroup M3 and mutation at 16129 designates haplogroup M1.
This is what you see if you visit phylotre- the official naming site.
Is this what you questioned?
Clearly Phylotree is unreliable and based on Euroocentrism and white supremacy. This is obvious given the way R-M173 in Africa was changed into V88, and 207 in Africa was changed into V45. All of these efforts are simply misdirection aimed at containing Negroes in Africa, and separating Dravidian speakers from Africans.
Researchers maintain that the Eastern African HVS-I signature motifs are 16,129, 16,189, 16,249, and 16,311. This motiff is found in Indian M1, so it can not be changed into M3, because M3 already existed.
.
.
The presence of one transition, 126, in Indian M1, does not deny its existence as M1, since it is the only M haplogroup outside of Africa that carries the entire M1 motiff. Luckily, we have the original paper that proves that M1 in India exist.
.
quote:
Originally posted by Mike111:
^So with the physical features debacle behind him, the Albino man moved on to genetics, and declared that Haplogroup "R" was HIS and his ALONE genetic source!
I BE DIFFERENT AND UNIQUE, SAYETH THE ALBINO MAN!
Anyone remember this?
King Tut Was a European!!!
DNA tests show that King Tut’s Y-DNA matches that of modern day Western Europeans. The lineup is with R1b.
Okay, lets look at the "WHITE MAN" haplogroup.

Right away we see a problem for the White man, turns out that "R" is NOT a White mans haplogroup after all.
So what does the White man do?
Why he digs DEEPER, clearly his intent is to keep digging until he finds a SUB-haplogroup that is exclusively White.
LETS SEE HOW HE DID.
(Much of the text below serves only as Humor - Sounds like Lioness bullsh1t).
Haplogroup R1b
In human genetics, Haplogroup R1b is the most frequently occurring Y-chromosome haplogroup in Western Europe, parts of central Eurasia (for example Bashkortostan), and in parts of sub-Saharan Central Africa (for example around Chad and Cameroon). R1b is also present at lower frequencies throughout Eastern Europe, Western Asia, Central Asia, and parts of South Asia and North Africa. Due to European emigration it also reaches high frequencies in the Americas and Australia. While Western Europe is dominated by the R1b1a2 (R-M269) branch of R1b, the Chadic-speaking area in Africa is dominated by the branch known as R1b1c (R-V88). These represent two very successful "twigs" on a much bigger "family tree."
R1b1c is found in northern Cameroon in west central Africa at a very high frequency, where it is considered to be caused by a pre-Islamic movement of people from Eurasia.
Suggestive results from other studies which did not test for the full range of new markers discovered by Cruciani et al. have also been reported, which might be in R-V88.
Wood et al. reported high frequencies of men who were P25 positive and M269 negative, amongst the same north Cameroon area where Cruciani et al. reported high R-V88 levels. However they also found such cases amongst 3% (1/32) of Fante from Ghana, 9% (1/11) of Bassa from southern Cameroon, 4% (1/24) of Herero from Namibia, 5% (1/22) of Ambo from Namibia, 4% (4/92) of Egyptians, and 4% (1/28) of Tunisians.
Luis et al. found the following cases of men M173 positive (R1), but negative for M73 (R1b1b1), M269 (R1b1b2), M18 (R1b1a1, a clade with V88, M18 having been discovered before V88) and M17 (R1a1a): 1 of 121 Omanis, 3 of 147 Egyptians, 2 of 14 Bantu from southern Cameroon, and 1 of 69 Hutu from Rwanda.
Pereira et al. (2010) in a study of several Saharan Tuareg populations, found one third of 31 men tested from near Tanut in Niger to be in R1b.
R1b1c1 (R-M18)
R1b1c1 is a sub-clade of R-V88 which is defined by the presence of SNP marker M18. It has been found only at low frequencies in samples from Sardinia and Lebanon.
Historical note
The DNA tests that assisted in the identification of Czar Nicholas II of Russia found that he had haplogroup R1b.

quote:Mike you are on point. Europeans have changed the names of haplogroups or simply deny the African origin of haplogroups to pretend that hg E is the the sole African haplogroup.
Originally posted by Mike111:
quote:Lioness - if I were to take what you Albinos say seriously, then I would be just as fuched-up as you.
Originally posted by the lioness:
note: typing "he he he" and "ha ha ha" won't save you
Lets go over some of the bullsh1t the Albino lying idiots of the study "Melungeons, A Multi-Ethnic Population" i.e.
Roberta J. Estes, Jack H. Goins, Penny Ferguson, Janet Lewis Crain, said.
Haplogroup R1b1b2 is of European origin (page 50)
Primary Gibson Group Ethnicity
This group is haplogroup R1b1b2ab5 (L21) known to be Celtic. This haplogroup suggests that
the Gibsons from this line who are documented to be "of color" do not carry that designation as a
result of the paternal Y-line. (page 53)
Gibson E1b1a Ethnicity
This group is haplogroup E1b1a of sub-Saharan African origin. The census and concealed
tithables records in Louisa County suggest that members of this genetic line were "of color"
which is supported by the haplogroup designation. (page 54).
Moore Ethnicity
This group of three individuals is haplogroup R1b1b2a1b, European, believed to be Celtic.
Historical records indicate that this Moore family was "of color", but the haplogroup indicates
that the paternal Y-line was not the source. (page 65)
The Albino point to all of that, was to say that ONLY those of haplogroup "E" had Black fathers because "R" was Whites ONLY. Clearly Albinos are once again attempting to deny their Albinohood by carving out some genetic "EXCLUSIVE" ground for themselves.
At this point it would be illuminating to look back at the Albinos first attempts at this.
Before genetics, the Albinos tried to use facial features to distinguish themselves from normal humans.
Only Whites had high nose bridges, thin lips and straight hair.
When it was pointed out to them, that Blacks had those features even more extreme than they did:

Then they said that ONLY us Whites have multi-colored hair, and eyes. (all indicators of Albinism) silly Albinos!

There are a couple of threads here on ES with more pictures, so I won't bother.
So now, with all of his Albino denying bullsh1t debunked, the Albino man moved on to genetics, to try and convince himself that he is not an Albino.
(See what I mean by He,he,he: and Ha,ha,ha: Lioness)?
Next we shall see what the Albino man actually learned about himself - But that didn't and won't, stop him from LYING!
quote:
Originally posted by Mike111:
Haplogroup R1a
R1a and R1a1a are believed to have originated somewhere within Eurasia, most likely in the area from Eastern Europe to South Asia. Several recent studies have proposed that South Asia is the most likely region of origin. But on the other hand, as will be discussed below, some researchers continue to treat modern Indian R1a as being largely due to immigration from the Central Eurasian steppes or Southwestern Asia.
R1a has been found in high frequency at both the eastern and western ends of its core range, for example in India and Tajikistan on the one hand, and Poland on the other. Throughout all of these regions, R1a is dominated by the R1a1a (R-M17 or R-M198) sub-clade.
In South Asia R1a1a has often been observed with high frequency in a number of demographic groups. The main two subclades of R1a1a are R1a1a* and R1a1a7. R1a1a7 is positive for M458 an SNP that separate it from the rest of R1a1a. It is significant because M458 is a European marker and the epicenter is Poland. M458 marker is rare in India.
In India, high percentage of this haplogroup is observed in West Bengal Brahmins (72%) to the east, Konkanastha Brahmins (48%) to the west, Khatris (67%) in north and Iyenger Brahmins (31%) of south. It has also been found in several South Indian Dravidian-speaking Adivasis including the Chenchu (26%) and the Valmikis of Andhra Pradesh and the Kallar of Tamil Nadu suggesting that M17 is widespread in Tribal Southern Indians.
Besides these, studies show high percentages in regionally diverse groups such as Manipuris (50%) to the extreme North East and in Punjab (47%) to the extreme North West.
In Pakistan it is found at 71% among the Mohanna tribe in Sindh province to the south and 46% among the Baltis of Gilgit-Baltistan to the north. While 13% of Sinhalese of Sri Lanka were found to be R1a1a (R-M17) positive.
Hindus of Terai region of Nepal show it at 69%.
In Afghanistan, R1a1a (R-M17) is found at 51.02% among the Pashtuns (the largest ethnic group in Afghanistan) and 30.36% among the Tajiks, but it is less frequent among the Hazaras (6.67%) and the Turkic-speaking Uzbeks (17.65%).
Europe
R1a1 among others European haplogrupes
In Europe, R1a, again almost entirely in the R1a1a sub-clade, is found at highest levels among peoples of Eastern European descent (Sorbs, Poles, Russians and Ukrainians; 50 to 65%). In the Baltic countries R1a frequencies decrease from Lithuania (45%) to Estonia (around 30%). Levels in Hungarians have been noted between 20 and 60%.
There is a significant presence in peoples of Scandinavian descent, with highest levels in Norway and Iceland, where between 20 and 30% of men are in R1a1a. Vikings and Normans may have also carried the R1a1a lineage westward; accounting for at least part of the small presence in the British Isles. In East Germany, where Haplogroup R1a reaches a peak frequency in Rostock at a percentage of 31.3%, it averages between 20%-30%.
Haplogroup R1a1a was found at elevated levels amongst a sample of the Israeli population who self-designated themselves as Ashkenazi Jews, possibly reflecting gene flow into Ashkenazi populations from surrounding Eastern European populations, over a course of centuries. This haplogroup finding was apparently consistent with the latest SNP microarray analysis which argued that up to 55 percent of the modern Ashkenazi genome is specifically traceable to Europe. Ashkenazim were found to have a significantly higher frequency of the R-M17 haplogroup Behar reported R-M17 to be the dominant haplogroup in Ashkenazi Levites (52%), although rare in Ashkenazi Cohanim (1.3%) and Israelites (4%).
In Southern Europe R1a1a is not common amongst the general population, but it is widespread in certain areas. Significant levels have been found in pockets, such as in the Pas Valley in Northern Spain, areas of Venice, and Calabria in Italy. The Balkans shows lower frequencies, and significant variation between areas, for example >30% in Slovenia, Croatia and Greek Macedonia, but <10% in Albania, Kosovo and parts of Greece.
The remains of a father and his two sons, from an archaeological site discovered in 2005 near Eulau "in Saxony-Anhalt, Germany" and dated to about 2600 BCE, tested positive for the Y-SNP marker SRY10831.2. The R1a1 clade was thus present in Europe at least 4600 years ago, in association with one site of the widespread Corded Ware culture.
Central and Northern Asia
R1a1a frequencies are patchy in Central Asia. This variation is possibly a consequence of population bottlenecks in isolated areas and the movements of Scythians in ancient times and later the Turco-Mongols.
High frequencies of R1a1a "R-M17 or R-M198; 50 to 70%" are found among the Ishkashimis, Khujand Tajiks, Panjakent Tajiks, Turkic-speaking Kyrgyzs, and in several peoples of Russia's Altai Republic, but frequencies are relatively lower "16 to 25%" among the Dushanbe Tajiks, Samarkand Tajiks, Yaghnobis and Shughnis.
Although levels are comparatively low amongst some Turkic-speaking groups e.g. Turks, Azeris, Kazakhs, Yakuts, levels are high "19 to 28%" in certain Turkic or Mongolic-speaking groups of Northwestern China, such as the Bonan, Dongxiang, Salar, and Uyghurs.
In Eastern Siberia, R1a1a is found among certain indigenous ethnic groups including Kamchatkans and Chukotkans, and peaking in Itel'man at 22%.
Middle East and Caucasus
R1a1a has been found in various forms, in most parts of Western Asia, in widely varying concentrations, from almost no presence in areas such as Jordan, to much higher levels in parts of Kuwait, Turkey and Iran.
The Shimar "Shammar" Bedouin tribe in Kuwait show the highest frequency in the Middle East at 43%.
Wells et al. (2001), noted that in the western part of the country, Iranians show low R1a1a levels, while males of eastern parts of Iran carried up to 35% R1a. Nasidze et al. (2004) found R1a in approximately 20% of Iranian males from the cities of Tehran and Isfahan. Regueiro et al. (2006), in a study of Iran, noted much higher frequencies in the south than the north.
Turkey also shows high but unevenly distributed R1a levels amongst some sub-populations. For example Nasidze et al. (2005) found relatively high levels amongst two Kurdish groups of Turkey, the Kurmanji (13%) and Zazaki (26%).
Further to the north of these Middle Eastern regions on the other hand, R1a levels start to increase in the Caucasus, once again in an uneven way. Several populations studied have shown no sign of R1a, while highest levels so far discovered in the region appears to belong to speakers of the Karachay-Balkar language amongst whom about one quarter of men tested so far are in haplogroup R1a1a.
quote:
Originally posted by Mike111:
^So then, putting aside the very "IMAGINATIVE" conclusions of those "WHITE" researchers.
Lets us normal people look at the evidence without the bullsh1t.
IF THIS MAN:
AND THIS MAN:
WHO ARE BLACK, SHARE PRETTY MUCH THE EXACT SAME DNA AS THIS MAN:
WHO IS "WHITE"
THEN WHAT DOES THAT MEAN????
------------------------------------------------------------------------------------------------------------------------------------------------------
Oh wait, I seem to remember running into this same problem a while back.
As I recall, these children also shared the exact same DNA, yet one was White and the others were Black.

.
Hmmm, what did that mean again?
Oh yes, now I remember!
THE WHITE ONE WAS AN ALBINO!

 [/URL]
[/URL]
quote:DAMNNNN - YOU'RE STUPID!
Originally posted by the lioness,:
notice the caption on the chart
black circles don't represent anything


quote:Only in your fevered imagination. Who else has published these claims?
Originally posted by Clyde Winters:
Africans carry haplogroup R1a.
In India the Dravidian people carry the R1a haplogroup The Dravidian people of India originally lived in Middle Africa and belonged to the Proto-Saharan Civilization.
The Proto-Saharan civilization was situated in the Proto-Sahara, which includes Cameroon.
.
.
In Cameroon we find carriers of R1a.
In addition to carriers of R1a in Cameroon; the Dravidian languages are still spoken today in Cameroon see: https://www.youtube.com/watch?v=vWyAYGlFZjkhttps://www.youtube.com/watch?v=vWyAYGlFZjk
quote:Good catch! doctoring the evidence disguised by lots of spam postings
Originally posted by the lioness,:

This is a misleading wikipedia chart on R1, the two branches R1a and R1b
the black circles are blank they don't represent anything by themseleves. Mike added the yellow caption "R1 Black"(scam)
The chart does not come from a science source reference
Notice how those black circles don't even cover the below Cruciani chart coastal WA positions
[/URL]
Cruciani et al (2010) has renamed the R*-
M173 (R P-25) in Africa V88. The TMRCA of V88 was
9200-5600 kya (Cruciani et al, 2010). Specific alleles distinguish this branch
https://en.wikipedia.org/wiki/Haplogroup_R1
notice the caption on the chart
black circles don't represent anything
quote:Yeah, thanks for the reply.
Originally posted by Quetzalcoatl:
quote:In 1999 potential confusion arose because 2 papers were published that called certain sequences M1. One had to change names, and Phylotree designated that the one carrying M16126 would now be named M3.
Originally posted by Troll Patrol # Ish Gebor:
quote:So, M1 became M3 somewhere in 1999.
Originally posted by Quetzalcoatl:
quote:Yes,
Originally posted by Troll Patrol # Ish Gebor:
[qb] @ Quetzalcoatl,
Are these the papers you are referring at, when you spoke of the M1-M3 transposition?
4. Kivisild T, Kaldma K, Metspalu M, Parik J, Papiha S, Villems R. 1999. The place of the Indian mtDNA variants in the global network of maternal lineages and the peopling of the Old World. In Deka R, Papiha SS. eds Genomic Diversity Kluwer Academic/Plenum Publishers New York p 135–152.
5. Quintana-Murci L, Semino O, Bandelt H-J., Passarino G, McElreavey K, Santachiara-Benerecetti AS. 1999. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet 23: 437 – 441.
The question I'm having here is, what is the "new" M = M1 reference?
M1 16129 16189 16249 16311
M3 16126
Thus, it is absolutely clear that, at least since the year 2000, mutation at 16126 has designated haplogroup M3 and mutation at 16129 designates haplogroup M1.
This is what you see if you visit phylotre- the official naming site.
Is this what you questioned?
quote:
Originally posted by Quetzalcoatl:
quote:Good catch! doctoring the evidence disguised by lots of spam postings
Originally posted by the lioness,:

This is a misleading wikipedia chart on R1, the two branches R1a and R1b
the black circles are blank they don't represent anything by themseleves. Mike added the yellow caption "R1 Black"(scam)
The chart does not come from a science source reference
Notice how those black circles don't even cover the below Cruciani chart coastal WA positions
[/URL]
Cruciani et al (2010) has renamed the R*-
M173 (R P-25) in Africa V88. The TMRCA of V88 was
9200-5600 kya (Cruciani et al, 2010). Specific alleles distinguish this branch
https://en.wikipedia.org/wiki/Haplogroup_R1
notice the caption on the chart
black circles don't represent anything

quote:--Hong Shi et al. 2008:
The Y chromosome Alu polymorphism (YAP, also called M1) defines the deep-rooted haplogroup D/E of the global Y-chromosome phylogeny [1]. This D/E haplogroup is further branched into three sub-haplogroups DE*, D and E (Figure 1). The distribution of the D/E haplogroup is highly regional, and the three subgroups are geographically restricted to certain areas, therefore informative in tracing human prehistory (Table 1). The sub-haplogroup DE*, presumably the most ancient lineage of the D/E haplogroup was only found in Africans from Nigeria [2], supporting the "Out of Africa" hypothesis about modern human origin. The sub-haplogroup E (E-M40), defined by M40/SRY4064 and M96, was also suggested originated in Africa [3-6], and later dispersed to Middle East and Europe about 20,000 years ago [3,4]. Interestingly, the sub-haplogroup D defined by M174 (D-M174) is East Asian specific with abundant appearance in Tibetan and Japanese (30–40%), but rare in most of other East Asian populations and populations from regions bordering East Asia (Central Asia, North Asia and Middle East) (usually less than 5%) [5-7]. Under D-M174, Japanese belongs to a separate sub-lineage defined by several mutations (e.g. M55, M57 and M64 etc.), which is different from those in Tibetans implicating relatively deep divergence between them [1]. The fragmented distribution of D-M174 in East Asia seems not consistent with the pattern of other East Asian specific lineages, i.e. O3-M122, O1-M119 and O2-M95 under haplogroup O [8,9].
quote:Haplogroup DE* in Guinea-Bissau:
Further refinement awaits the finding of new markers especially within paragroup E3a*-M2. The microsatellite profile of the DE* individual is one mutational step away from the allelic state described for Nigerians (DYS390*21, DYS388 not tested; [37], therefore suggesting a common ancestry but not elucidating the phylogenetics.
quote:Haplogroup DE* in Nigerians:
There has been considerable debate on the geographic origin of the human Y chromosome Alu polymor- phism (YAP). Here we report a new, very rare deep-rooting haplogroup within the YAP clade, together with data on other deep-rooting YAP clades. The new haplogroup, found so far in only five Nigerians, is the least-derived YAP haplogroup according to currently known binary markers. However, because the interior branching order of the Y chromosome genealogical tree remains unknown, it is impossible to impute the origin of the YAP clade with certainty. We discuss the problems presented by rare deep-rooting lineages for Y chromosome phylogeography.
quote:--Fulvio Cruciani et al.
‘‘Out of Africa’’ haplogroups.
All Y-clades that are not exclusively African belong to the macro-haplogroup CT, which is defined by mutations M168, M294 and P9.1 [14,31] and is subdivided into two major clades, DE and CF [1,14].
In a recent study [16], sequencing of two chromosomes belonging to haplogroups C and R, led to the identification of 25 new mutations, eleven of which were in the C-chromosome and seven in the R-chromosome.
Here, the seven mutations which were found to be shared by chromosomes of haplogroups C and R [16], were also found to be present in one DE sample (sample 33 in Table S1), and positioned at the root of macro-haplogroup CT (Figure 1 and Figure S1)
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3492319/figure/pone-0049170-g001/
Figure S1
Structure of the macro-haplogroup CT. For details on mutations see legend to Figure 1. Dashed lines indicate putative branchings (no positive control available). The position of V248 (haplogroup C2) and V87 (haplogroup C3) compared to mutations that define internal branches was not determined. Note that mutations V45, V69 and V88 have been previously mapped (Cruciani et al. 2010; Eur J Hum Genet 18∶800–807).
(TIF)
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3492319/bin/pone.0049170.s001.tif
quote:Use of Y Chromosome and Mitochondrial DNA Population Structure in Tracing Human Migrations
"haplogroup CF and DE molecular ancestors first evolved inside Africa and subsequently contributed as Y chromosome founders to pioneering migrations that successfully colonized Asia. While not proof, the DE and CF bifurcation (Figure 8d ) is consistent with independent colonization impulses possibly occurring in a short time interval."
quote:The Mal'ta boy was Khoisan. His ancestors were the first anatomically modern humans.
Originally posted by Troll Patrol # Ish Gebor:
The Mal'ta boy didn't fell from the sky, onto Siberia near Lake Baikal?
quote:
“The genetic profile of the Paleoamericans, fails to correspond to that of contemporary Native Americans in the United States. Interestingly, the North American Paleoamerican DNA profile matches minor haplogroups predominately found in South America (Balter, 2015). Much of the DNA for Luzia the 12,000 year old skeleton from Brazil is corrupted, but researchers have recovered aDNA from Naia (Chatters et al., 2014) of Mexico, and the Anzick boy (Balter, 2015; Estes, 2015). The Anzick boy skeleton was found in Montana. This Paleoamerican belonged to the Clovis Culture, the same as Kennewick Man (Chatters, 1999; Chatters et al., 2014). The Anzick boy belonged to mtDNA M or D1, and y-chromosome R1 (Estes, 2015). Scientists have also recovered the DNA fromNaia (Chatters et al., 2014; Kumar, 2014). Naia belonged to haplogroup D1, which is a descendant of the M haplogroup (Chatters et al., 2014).
Researchers have also recovered the aDNA of the ancient Europeans (Balter, 2015). The TMRCA of the paleoamericans were the Khoisan people. The Khoisan was the Cro-Magnon people of Europe (Winters, 2008, 2011). They were the first amh to enter western Eurasia (Winters, 2011). The Khoisan introduced haplogroup M to western Eurasia (Winters, 2011, 2014).
The first Europeans like the paleoamericans were dark skinned (Winters, 2014). The aDNA of the first Europeans comes from the Ust’Ishim skeleton from Siberia (Blater, 2015). Ust’Ishim man carried the male lineage R1 (Balter, 2015; Immanuel, 2014a, 2014b). The mtDNA of Ust’Ishim belonged to haplogroup U (Balter, 2015). The R1 haplogroup was also carried by the Mal’ta boy in Western Eurasia (Blater, 2015; Immanuel, 2014a, 2014b). The Mal’ta boy also belonged to mtDNA haplogroup U (Balter, 2015; Immanuel, 2014a, 2014b). The U haplogroup was part of the M macrohaplogroup. The Khoisan carry haplogroupsL3(M, N).
Prior to the Khoisan crossing the Straits of Gibraltar to reach Iberia, they probably stopped in West Africa (Winters, 2014). The basal L3(M) motif in West Africa is characterized by the Ddel site np 10,394 and Alul site np 10,397 which is associated with AF-24, a haplotype of haplogroup LOd (Winters, 2010). Granted L3 and L2 are not as old as LOd, but Gonder et al., (2006), provides an early date for, L3(M, N) 94.3kya. The South African Khoisan (SAK) carry L1c, L1, L2, L3 M, N dates to 142.3 kya; the Hadza are L2a, L2, L3, M, N, dates to 96.7 kya (Gonder et al., 2006).
The origin dates for L1, L2, L3(M, N) make the haplogroups old enough for the Khoisan to have taken haplogroup M to West Africa, where we find L3, L2 and LOd and thence to Iberia (Winters, 2011). It is interesting to note that LO haplogroups are primarily found among Khoisan and West Africans (Winters, 2011). This shows that at some point in prehistory the Khoisan had migrated into West Africa. The major M haplogroup in Africa is M1(Winters, 2010). The M1 macrohaplogroup is found throughout Africa and Asia. But the basal M1 lineage has not been found outside Africa (Sun et al., 2006). However, on the basis of currently available FGS sequences, M1 markers have been found in the D4a branch of Haplogroup D, the most widespread branch of M1 in East Asia (Fucharoen et al., 2001; Yao et al., 2002). These transitions are recurrent in M1 and D4 (Gondor et al., 2006; Winters, 2010).”
See: http://www.cibtech.org/J-Microbiology/PUBLICATIONS/2015/Vol-4-No-1/03-CJM-004-CLYDE-AFRICAN-DNA.pdf
quote:This is my response to this article.
Originally posted by the lioness,:
http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0007447
2009
Updating Phylogeny of Mitochondrial DNA Macrohaplogroup M in India: Dispersal of Modern Human in South Asian Corridor
Adimoolam Chandrasekar et al
Published: October 13, 2009
DOI: 10.1371/journal.pone.000744
Abstract
To construct maternal phylogeny and prehistoric dispersals of modern human being in the Indian sub continent, a diverse subset of 641 complete mitochondrial DNA (mtDNA) genomes belonging to macrohaplogroup M was chosen from a total collection of 2,783 control-region sequences, sampled from 26 selected tribal populations of India. On the basis of complete mtDNA sequencing, we identified 12 new haplogroups - M53 to M64; redefined/ascertained and characterized haplogroups M2, M3, M4, M5, M6, M8′C′Z, M9, M10, M11, M12-G, D, M18, M30, M33, M35, M37, M38, M39, M40, M41, M43, M45 and M49, which were previously described by control and/or coding-region polymorphisms. Our results indicate that the mtDNA lineages reported in the present study (except East Asian lineages M8′C′Z, M9, M10, M11, M12-G, D ) are restricted to Indian region.The deep rooted lineages of macrohaplogroup ‘M’ suggest in-situ origin of these haplogroups in India. Most of these deep rooting lineages are represented by multiple ethnic/linguist groups of India. Hierarchical analysis of molecular variation (AMOVA) shows substantial subdivisions among the tribes of India (Fst = 0.16164). The current Indian mtDNA gene pool was shaped by the initial settlers and was galvanized by minor events of gene flow from the east and west to the restricted zones. Northeast Indian mtDNA pool harbors region specific lineages, other Indian lineages and East Asian lineages. We also suggest the establishment of an East Asian gene in North East India through admixture rather than replacement.
Origin of Macrohaplogroup M
L3 lineages other than M and N are absent in India and among non-African mitochondria in general [2]–[3], [49]. M, N and R haplogroups of mtDNA have no indication of an African origin. However, it is proposed that the origin of haplogroup M is in Africa [34], in view of its high frequency in Ethiopia. But in 2006, by [35] demonstrated that the presence of M1 and U6 in Africa is due to a back migration. Sequencing of 81 entire human mitochondrial DNAs belonging to haplogroups M1 and U6 revealed that these predominantly North African Clades arose in Southwestern Asia and moved together to Africa about 40,000 to 45,000 years ago. Only some sub-sets of M1a (with an estimated coalescence time of 28.8±4.9ky), U6a2 (with an estimated coalescence time of 24.0±7.3ky), and U6d (with an estimated coalescence time of 20.6±7.3ky) diffused to East and North Africa through the Levant, leaving the origin of macrohaplogroup M unresolved. Haplogroup M has been found ubiquitous in India, although its frequency is somewhat higher in southern Indian populations than in northern Indian populations and to a large extent autochthonous because neither the East nor the West Eurasian mtDNA pools include such lineages at notable frequencies [37], [58]. Our findings, (for example, deep time depth >50,000 years of western, central, southern and eastern Indian haplogroups M2, M38, M54, M58, M33, M6, M61, M62 and distribution of macrohaplogroup M) do not rule out the possibility of macrohaplogroup M arising in Indian population.
Migration routes of modern human
Recent mtDNA evidence on modern human out of Africa migration route suggests a single dispersal by a southern coastal route to India and further, to East Asia and Australia [17], [20], [22], [23], [66], [69]. The North Asian route could not get support from mtDNA due to the lack of basal M, R, N lineages in northern Asians, thereby ruling out the existence of a northern Asian route [29]–[30], [70]–[71]. Proven back migration of sub lineages of M and U into Africa [35], and the absence of L3 lineages or ancestral lineage for L3, M and N in India, leaves two issues unresolved: evidences for the southern route hypothesis from India and origin of M haplogroup. However, in the present study, the basal diversity (37 nodes) and founder ages (57,000–75,000 years) of macrohaplogroup M in India reveals initial settlement of African exodus in India. Our database also reveals evidences that Andaman islanders and Australians have ancestral maternal roots in India [24], [43].
In summary, the present study provides evidence that several Indian mtDNA M lineages are deep rooted and in situ origin. In North East India the coalescent time of East Asian lineages dates back to Last Glacial Maximum (LGM). Further, the combination of virtually all previously reported lineages from South and East Asia and our newly produced Indian complete mtDNA sequences have helped to define several novel (sub) haplogroups. The present work further ascertained previously reported haplogroups, and refined the phylogenetic tree of South Asia. This updated phylogenetic tree provides an essential reference guide for diseases, anthropological and forensic studies among Asian populations.
quote:
- Macrohaplogroup M Did not Originate in India
Posted by Clyde98 on 21 Aug 2011 at 06:11 GMT
by
Clyde Winters
Chandrasekar et al provide a good discussion of the phylogeny of Indian macrohaplogroup M. Although they claimed to have examined all of the M haplogroups in India they failed to discuss haplogroup M1, which is also found in India [3-4]. This failure to discuss all the M lineages in India cast doubt on the conclusions of the authors of this study.
The researchers argue that the M macrohaplogroup in India developed in situ. They base this claim on the research of Gonzalez et al [1].
Chandrasekar et al maintain that the research in [1] indicates that M1 probably originated in Southwest Asia and through a back migration hg M and U6 returned to Africa. This is false hg M probably originated in Africa, not Asia [3].
Chandrasekar et al claim that hg M arose in Southwest Asia 40kya. This date is ludicrous because Neanderthals lived in that region at this time. The only anatomically modern humans in Western Eurasia at this time were Cro-Magnon man who carried haplogroup N.
To estimate the coalescence age of haplogroup M1 Gonzalez et al [1] analyzed 13 complete sequences of haplogroup M1.
Gonzalez et al claims that the M1c lineage is the oldest M1 subclade based on the coalescence age estimation of the M1 subgroup: M1a (16756 +-5997), M1b (10155 +-3590) and M1c (19040+-4916). This makes M1a and M1b the youngest clades.
The available sample for M1c was complete sequences from individuals found in Jordan, Senegal, and Spain. The small data set make a precise estimation of the errors in the data uncertain.
The limited sample for M1c makes it difficult to effectively quantify the estimation error for the data, since error increases from level to level in models possessing a hierarchical structure.
The small sample size makes the confidence intervals overlap. This calls into question the conclusions of Gonzalez et al [1] in relation to the ages of hg M1 despite the differing levels of hierarchy.
In addition to the evidence of the coalescence age estimation in support of the antiquity of M1c, Gonzalez et al believe the presence of M1c among Jordanians is an important indicator for the ancient origin of this clade. The evidence of M1c in Jordan, does not really add to the hypothesis that M1c is the oldest clade because the presence of this clade in the Middle East can be explained by the thousands of West Africans who have taken the hajj to Mecca, and remained in the Middle East, instead of returning to West Africa.
The Valencia sample can also be explained by the history of Islam. There is a direct link between Senegal and Yusuf ibn Tashufin. Yusuf founded the Almoravids. The Almoravid empire extended from Senegal to Spain [2].
This link comes from the fact that many of the followers of Tashufin came from the ribats or ‘religious schools’ he had established in northern Senegal. Troops from these ribats formed the backbone of Tashufin’s army when he invaded Spain in 1086[2]. These African Muslims ruled much of Spain until 1492. Since M1c is presently found in Senegal, the carrier of M1c reported by Gonzalez et al in Valencia may be a descendent of these African Almoravids that ruled Spain for over 700 years
Sub-Saharan Africans probably spread hg M1c to Eurasia. Gonzalez et al reported that the carriers of the M1c subset were from Jordan, Senegal and Valencia [1]. It was revealed above that 1) many of the Muslim troops in Tashufin’s army that conquered Spain in 1086 AD, came from Senegal; and 2) many West Africans after taking the Hajj, visited Jerusalem and settled in the Middle East. Even if we eliminate the Jordan sample, the evidence from Valencia and Senegal gives a 67% probability that M1c originated in Senegal, not Asia or North Africa because of the historical presence of Sub-Saharan Africans in both areas . This provides support for an African origin of M1.
Chandrasekar et al claim that India is the only region where there is a variety of M subclades is also false. In Africa, for example in addition to M1, we also find haplogroups M3,M30 and M33.
Chandrasekar et al claims that there is no influence of African haplogroups in India. The presence of M1 among South Indian Dravidian speakers make it clear that African mtDNA is found in India [1][3]. This along with African y-chromosomes and African HLA among Dravidian tribal groups indicate a recent African influence among South Indians [4-6]. This is not surprising since Dravidian speakers formerly belonged to the C-Group culture of Nubia, and only entered India 5kya [4].
The distribution of continental African populations carrying M haplogroups favors Africa as the place of origin for this macrohaplogroup instead of India. The population distributions for the M macrohaplogroup in Africa make it clear that haplogroup M originated in Africa, not Asia or North Africa.
Reference:
1. Gonzalez , A. Jose M Larruga , Khaled K Abu-Amero , Yufei Shi , Jose Pestano and Vicente M Cabrera. (2007).Mitochondrial lineage M1 traces an early human backflow to Africa, BMC Genomics , 8:223 doi:10.1186/1471-2164-8-223. Retrieved on 9/15/2010 http://www.biomedcentral....
2. Bovill,E.W. (1970). The Golden trade of the Moors. London: Oxford University Press.
3. Winters,C. (2010). The African Origin of the M1 Haplogroup Introduction. Current Research Journal of Biological Sciences. Retrieved 8/20/2011 http://olmec98.net/afro_m...
4. Winters, C.(2007). Did the Dravidian Speakers Originate in Africa? BioEssays,27(5):497-498.
5.Winters, C. (2010). Y-chromosome evidence of an African origin of Dravidian Agriculture. Int J Genet & Molec Bio, 2(3):030-033. Retrieved 6/4/2010 at: http://www.academicjourna...
6. Winters, C. (2010). 9bp and the Relationship Between African and Dravidian Speakers. Current Research Journal of Biological Sciences 2(4): 229-231. http://maxwellsci.com/pri...
See: http://www.plosone.org/annotation/listThread.action?root=6633
.
quote:Try to find one peer review article that is not written by Clyde Winters that even mentions Dravidians and Nubian C group in the same article
Originally posted by Troll Patrol # Ish Gebor:
Yeah, thanks for the reply.
This was posted by Clyde, do you agree?
The majority of M clades in Asia are carried by Dravidian people, Dravidian people originated in Africa and belonged to the C-Group people.
quote:stop making up stuff, thanks
Originally posted by Clyde Winters:
The Mal'ta boy was Khoisan.
quote:Mainly spam. Your comment. continues your delusion that there is M1 in India. Either it is mental block, or you are so invested in this falsehood, that losing it will endanger all your work because the Dravidians are the key for your extension of "proto-Saharans" to other parts of the world. M1 and M3 are two things 1)labels for a series of mutations and 2) designations of the haplogroups people have. One can change role one as nomenclature conflicts arise without making role two disappear. You can claim that haplogroup M1 is present in India till the cows come home BUT There are no people in India with a haplogroup with SNPs (T195C! G6446A T6680C C12403T A12950c G16129A! T16189C! T16249C T16311C) i.e. the real M1. on the other hand there are millions in India with haplotype containing the SNPs (T482C T16126C ) i.e. M3.
Originally posted by Clyde Winters:
quote:This is my response to this article.
Originally posted by the lioness,:
http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0007447
2009
Updating Phylogeny of Mitochondrial DNA Macrohaplogroup M in India: Dispersal of Modern Human in South Asian Corridor
Adimoolam Chandrasekar et al
Published: October 13, 2009
DOI: 10.1371/journal.pone.000744
Abstract
To construct maternal phylogeny and prehistoric dispersals of modern human being in the Indian sub continent, a diverse subset of 641 complete mitochondrial DNA (mtDNA) genomes belonging to macrohaplogroup M was chosen from a total collection of 2,783 control-region sequences, sampled from 26 selected tribal populations of India. On the basis of complete mtDNA sequencing, we identified 12 new haplogroups - M53 to M64; redefined/ascertained and characterized haplogroups M2, M3, M4, M5, M6, M8′C′Z, M9, M10, M11, M12-G, D, M18, M30, M33, M35, M37, M38, M39, M40, M41, M43, M45 and M49, which were previously described by control and/or coding-region polymorphisms. Our results indicate that the mtDNA lineages reported in the present study (except East Asian lineages M8′C′Z, M9, M10, M11, M12-G, D ) are restricted to Indian region.The deep rooted lineages of macrohaplogroup ‘M’ suggest in-situ origin of these haplogroups in India. Most of these deep rooting lineages are represented by multiple ethnic/linguist groups of India. Hierarchical analysis of molecular variation (AMOVA) shows substantial subdivisions among the tribes of India (Fst = 0.16164). The current Indian mtDNA gene pool was shaped by the initial settlers and was galvanized by minor events of gene flow from the east and west to the restricted zones. Northeast Indian mtDNA pool harbors region specific lineages, other Indian lineages and East Asian lineages. We also suggest the establishment of an East Asian gene in North East India through admixture rather than replacement.
Origin of Macrohaplogroup M
L3 lineages other than M and N are absent in India and among non-African mitochondria in general [2]–[3], [49]. M, N and R haplogroups of mtDNA have no indication of an African origin. However, it is proposed that the origin of haplogroup M is in Africa [34], in view of its high frequency in Ethiopia. But in 2006, by [35] demonstrated that the presence of M1 and U6 in Africa is due to a back migration. Sequencing of 81 entire human mitochondrial DNAs belonging to haplogroups M1 and U6 revealed that these predominantly North African Clades arose in Southwestern Asia and moved together to Africa about 40,000 to 45,000 years ago. Only some sub-sets of M1a (with an estimated coalescence time of 28.8±4.9ky), U6a2 (with an estimated coalescence time of 24.0±7.3ky), and U6d (with an estimated coalescence time of 20.6±7.3ky) diffused to East and North Africa through the Levant, leaving the origin of macrohaplogroup M unresolved. Haplogroup M has been found ubiquitous in India, although its frequency is somewhat higher in southern Indian populations than in northern Indian populations and to a large extent autochthonous because neither the East nor the West Eurasian mtDNA pools include such lineages at notable frequencies [37], [58]. Our findings, (for example, deep time depth >50,000 years of western, central, southern and eastern Indian haplogroups M2, M38, M54, M58, M33, M6, M61, M62 and distribution of macrohaplogroup M) do not rule out the possibility of macrohaplogroup M arising in Indian population.
Migration routes of modern human
Recent mtDNA evidence on modern human out of Africa migration route suggests a single dispersal by a southern coastal route to India and further, to East Asia and Australia [17], [20], [22], [23], [66], [69]. The North Asian route could not get support from mtDNA due to the lack of basal M, R, N lineages in northern Asians, thereby ruling out the existence of a northern Asian route [29]–[30], [70]–[71]. Proven back migration of sub lineages of M and U into Africa [35], and the absence of L3 lineages or ancestral lineage for L3, M and N in India, leaves two issues unresolved: evidences for the southern route hypothesis from India and origin of M haplogroup. However, in the present study, the basal diversity (37 nodes) and founder ages (57,000–75,000 years) of macrohaplogroup M in India reveals initial settlement of African exodus in India. Our database also reveals evidences that Andaman islanders and Australians have ancestral maternal roots in India [24], [43].
In summary, the present study provides evidence that several Indian mtDNA M lineages are deep rooted and in situ origin. In North East India the coalescent time of East Asian lineages dates back to Last Glacial Maximum (LGM). Further, the combination of virtually all previously reported lineages from South and East Asia and our newly produced Indian complete mtDNA sequences have helped to define several novel (sub) haplogroups. The present work further ascertained previously reported haplogroups, and refined the phylogenetic tree of South Asia. This updated phylogenetic tree provides an essential reference guide for diseases, anthropological and forensic studies among Asian populations.
quote:
- Macrohaplogroup M Did not Originate in India
Posted by Clyde98 on 21 Aug 2011 at 06:11 GMT
by
Clyde Winters
Chandrasekar et al provide a good discussion of the phylogeny of Indian macrohaplogroup M. Although they claimed to have examined all of the M haplogroups in India they failed to discuss haplogroup M1, which is also found in India [3-4]. This failure to discuss all the M lineages in India cast doubt on the conclusions of the authors of this study.
The researchers argue that the M macrohaplogroup in India developed in situ. They base this claim on the research of Gonzalez et al [1].
Chandrasekar et al maintain that the research in [1] indicates that M1 probably originated in Southwest Asia and through a back migration hg M and U6 returned to Africa. This is false hg M probably originated in Africa, not Asia [3].
Chandrasekar et al claim that hg M arose in Southwest Asia 40kya. This date is ludicrous because Neanderthals lived in that region at this time. The only anatomically modern humans in Western Eurasia at this time were Cro-Magnon man who carried haplogroup N.
To estimate the coalescence age of haplogroup M1 Gonzalez et al [1] analyzed 13 complete sequences of haplogroup M1.
Gonzalez et al claims that the M1c lineage is the oldest M1 subclade based on the coalescence age estimation of the M1 subgroup: M1a (16756 +-5997), M1b (10155 +-3590) and M1c (19040+-4916). This makes M1a and M1b the youngest clades.
The available sample for M1c was complete sequences from individuals found in Jordan, Senegal, and Spain. The small data set make a precise estimation of the errors in the data uncertain.
The limited sample for M1c makes it difficult to effectively quantify the estimation error for the data, since error increases from level to level in models possessing a hierarchical structure.
The small sample size makes the confidence intervals overlap. This calls into question the conclusions of Gonzalez et al [1] in relation to the ages of hg M1 despite the differing levels of hierarchy.
In addition to the evidence of the coalescence age estimation in support of the antiquity of M1c, Gonzalez et al believe the presence of M1c among Jordanians is an important indicator for the ancient origin of this clade. The evidence of M1c in Jordan, does not really add to the hypothesis that M1c is the oldest clade because the presence of this clade in the Middle East can be explained by the thousands of West Africans who have taken the hajj to Mecca, and remained in the Middle East, instead of returning to West Africa.
The Valencia sample can also be explained by the history of Islam. There is a direct link between Senegal and Yusuf ibn Tashufin. Yusuf founded the Almoravids. The Almoravid empire extended from Senegal to Spain [2].
This link comes from the fact that many of the followers of Tashufin came from the ribats or ‘religious schools’ he had established in northern Senegal. Troops from these ribats formed the backbone of Tashufin’s army when he invaded Spain in 1086[2]. These African Muslims ruled much of Spain until 1492. Since M1c is presently found in Senegal, the carrier of M1c reported by Gonzalez et al in Valencia may be a descendent of these African Almoravids that ruled Spain for over 700 years
Sub-Saharan Africans probably spread hg M1c to Eurasia. Gonzalez et al reported that the carriers of the M1c subset were from Jordan, Senegal and Valencia [1]. It was revealed above that 1) many of the Muslim troops in Tashufin’s army that conquered Spain in 1086 AD, came from Senegal; and 2) many West Africans after taking the Hajj, visited Jerusalem and settled in the Middle East. Even if we eliminate the Jordan sample, the evidence from Valencia and Senegal gives a 67% probability that M1c originated in Senegal, not Asia or North Africa because of the historical presence of Sub-Saharan Africans in both areas . This provides support for an African origin of M1.
Chandrasekar et al claim that India is the only region where there is a variety of M subclades is also false. In Africa, for example in addition to M1, we also find haplogroups M3,M30 and M33.
Chandrasekar et al claims that there is no influence of African haplogroups in India. The presence of M1 among South Indian Dravidian speakers make it clear that African mtDNA is found in India [1][3]. This along with African y-chromosomes and African HLA among Dravidian tribal groups indicate a recent African influence among South Indians [4-6]. This is not surprising since Dravidian speakers formerly belonged to the C-Group culture of Nubia, and only entered India 5kya [4].
The distribution of continental African populations carrying M haplogroups favors Africa as the place of origin for this macrohaplogroup instead of India. The population distributions for the M macrohaplogroup in Africa make it clear that haplogroup M originated in Africa, not Asia or North Africa.
Reference:
1. Gonzalez , A. Jose M Larruga , Khaled K Abu-Amero , Yufei Shi , Jose Pestano and Vicente M Cabrera. (2007).Mitochondrial lineage M1 traces an early human backflow to Africa, BMC Genomics , 8:223 doi:10.1186/1471-2164-8-223. Retrieved on 9/15/2010 http://www.biomedcentral....
2. Bovill,E.W. (1970). The Golden trade of the Moors. London: Oxford University Press.
3. Winters,C. (2010). The African Origin of the M1 Haplogroup Introduction. Current Research Journal of Biological Sciences. Retrieved 8/20/2011 http://olmec98.net/afro_m...
4. Winters, C.(2007). Did the Dravidian Speakers Originate in Africa? BioEssays,27(5):497-498.
5.Winters, C. (2010). Y-chromosome evidence of an African origin of Dravidian Agriculture. Int J Genet & Molec Bio, 2(3):030-033. Retrieved 6/4/2010 at: http://www.academicjourna...
6. Winters, C. (2010). 9bp and the Relationship Between African and Dravidian Speakers. Current Research Journal of Biological Sciences 2(4): 229-231. http://maxwellsci.com/pri...
See: http://www.plosone.org/annotation/listThread.action?root=6633
.
quote:
p. 688-689 A particular case in question is the origin of haplogroup M1, which is mainly found in Northeast Africa and the Near East (Quintana-Murci et al. 1999). Due to the fact that M1 bears variant nucleotides, for example, at site 16311 in common with haplogroup M4, at 16129 with M5, and at 16249 with haplogroup M34, it has been proposed that M1 might have some affinity with Indian M haplogroups (Roychoudhury et al. 2001). This inference, however, could not receive support from our complete sequencing information. Indeed, the reconstructed ancestral motifs of all Indian M haplogroups turned out to be devoid of those variations that characterized M1, that is, 6446, 6680, 12403, and 14110 (Maca-Meyer et al. 2001;Herrnstadt et al. 2002). Therefore, those common mutations in the control region rather reflect random parallel mutations. There is no evidence whatsoever that M1 originated in India.


quote:This argument is invalid. It implies that Phylotree promotes lies to maintain the status quo, and white supremacy.
Originally posted by Quetzalcoatl:
In 1999 potential confusion arose because 2 papers were published that called certain sequences M1. One had to change names, and Phylotree designated that the one carrying M16126 would now be named M3.
M1 16129 16189 16249 16311
M3 16126
Thus, it is absolutely clear that, at least since the year 2000, mutation at 16126 has designated haplogroup M3 and mutation at 16129 designates haplogroup M1.
This is what you see if you visit phylotre- the official naming site.
Is this what you questioned?

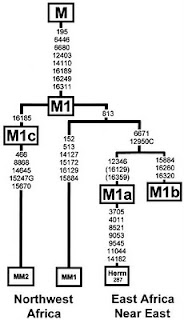
quote:There is nothing delusional in my proposition that M1 exist in India. Even Gonzalez (2007) admits that M1 was found in India and cites Kivisild et al (1999). Ana Gonzalez (2007) wrote “The central HVSI haplotype (16129–16189–16223–16249–16311) has been found only once in northwestern India [27]. Another possible Indian M1 candidate is the derived sequence: 16086–16129–16223–16249–16259–16311 [28]”.
Originally posted by Quetzalcoatl:
Mainly spam. Your comment. continues your delusion that there is M1 in India. Either it is mental block, or you are so invested in this falsehood, that losing it will endanger all your work because the Dravidians are the key for your extension of "proto-Saharans" to other parts of the world. M1 and M3 are two things 1)labels for a series of mutations and 2) designations of the haplogroups people have. One can change role one as nomenclature conflicts arise without making role two disappear. You can claim that haplogroup M1 is present in India till the cows come home BUT There are no people in India with a haplogroup with SNPs (T195C! G6446A T6680C C12403T A12950c G16129A! T16189C! T16249C T16311C) i.e. the real M1. on the other hand there are millions in India with haplotype containing the SNPs (T482C T16126C ) i.e. M3.
Sun, C. et al. “The Dazzling Array of Basal Branches in the mtDNA Macrohaplogroup M from India as Inferred from Complete Genomes,” Mol. Biol. Evol. 23(3):683–690.
quote:[/QB]
p. 688-689 A particular case in question is the origin of haplogroup M1, which is mainly found in Northeast Africa and the Near East (Quintana-Murci et al. 1999). Due to the fact that M1 bears variant nucleotides, for example, at site 16311 in common with haplogroup M4, at 16129 with M5, and at 16249 with haplogroup M34, it has been proposed that M1 might have some affinity with Indian M haplogroups (Roychoudhury et al. 2001). This inference, however, could not receive support from our complete sequencing information. Indeed, the reconstructed ancestral motifs of all Indian M haplogroups turned out to be devoid of those variations that characterized M1, that is, 6446, 6680, 12403, and 14110 (Maca-Meyer et al. 2001;Herrnstadt et al. 2002). Therefore, those common mutations in the control region rather reflect random parallel mutations. There is no evidence whatsoever that M1 originated in India.
quote:Of course he was, this is what I have been showing all this time.
Originally posted by Clyde Winters:
quote:The Mal'ta boy was Khoisan. His ancestors were the first anatomically modern humans.
Originally posted by Troll Patrol # Ish Gebor:
The Mal'ta boy didn't fell from the sky, onto Siberia near Lake Baikal?
“The genetic profile of the Paleoamericans, fails to correspond to that of contemporary Native Americans in the United States. Interestingly, the North American Paleoamerican DNA profile matches minor haplogroups predominately found in South America (Balter, 2015). Much of the DNA for Luzia the 12,000 year old skeleton from Brazil is corrupted, but researchers have recovered aDNA from Naia (Chatters et al., 2014) of Mexico, and the Anzick boy (Balter, 2015; Estes, 2015). The Anzick boy skeleton was found in Montana. This Paleoamerican belonged to the Clovis Culture, the same as Kennewick Man (Chatters, 1999; Chatters et al., 2014). The Anzick boy belonged to mtDNA M or D1, and y-chromosome R1 (Estes, 2015). Scientists have also recovered the DNA fromNaia (Chatters et al., 2014; Kumar, 2014). Naia belonged to haplogroup D1, which is a descendant of the M haplogroup (Chatters et al., 2014).
Researchers have also recovered the aDNA of the ancient Europeans (Balter, 2015). The TMRCA of the paleoamericans were the Khoisan people. The Khoisan was the Cro-Magnon people of Europe (Winters, 2008, 2011). They were the first amh to enter western Eurasia (Winters, 2011). The Khoisan introduced haplogroup M to western Eurasia (Winters, 2011, 2014).
The first Europeans like the paleoamericans were dark skinned (Winters, 2014). The aDNA of the first Europeans comes from the Ust’Ishim skeleton from Siberia (Blater, 2015). Ust’Ishim man carried the male lineage R1 (Balter, 2015; Immanuel, 2014a, 2014b). The mtDNA of Ust’Ishim belonged to haplogroup U (Balter, 2015). The R1 haplogroup was also carried by the Mal’ta boy in Western Eurasia (Blater, 2015; Immanuel, 2014a, 2014b). The Mal’ta boy also belonged to mtDNA haplogroup U (Balter, 2015; Immanuel, 2014a, 2014b). The U haplogroup was part of the M macrohaplogroup. The Khoisan carry haplogroupsL3(M, N).
Prior to the Khoisan crossing the Straits of Gibraltar to reach Iberia, they probably stopped in West Africa (Winters, 2014). The basal L3(M) motif in West Africa is characterized by the Ddel site np 10,394 and Alul site np 10,397 which is associated with AF-24, a haplotype of haplogroup LOd (Winters, 2010). Granted L3 and L2 are not as old as LOd, but Gonder et al., (2006), provides an early date for, L3(M, N) 94.3kya. The South African Khoisan (SAK) carry L1c, L1, L2, L3 M, N dates to 142.3 kya; the Hadza are L2a, L2, L3, M, N, dates to 96.7 kya (Gonder et al., 2006).
The origin dates for L1, L2, L3(M, N) make the haplogroups old enough for the Khoisan to have taken haplogroup M to West Africa, where we find L3, L2 and LOd and thence to Iberia (Winters, 2011). It is interesting to note that LO haplogroups are primarily found among Khoisan and West Africans (Winters, 2011). This shows that at some point in prehistory the Khoisan had migrated into West Africa. The major M haplogroup in Africa is M1(Winters, 2010). The M1 macrohaplogroup is found throughout Africa and Asia. But the basal M1 lineage has not been found outside Africa (Sun et al., 2006). However, on the basis of currently available FGS sequences, M1 markers have been found in the D4a branch of Haplogroup D, the most widespread branch of M1 in East Asia (Fucharoen et al., 2001; Yao et al., 2002). These transitions are recurrent in M1 and D4 (Gondor et al., 2006; Winters, 2010).”
See: http://www.cibtech.org/J-Microbiology/PUBLICATIONS/2015/Vol-4-No-1/03-CJM-004-CLYDE-AFRICAN-DNA.pdf
quote:As I asked you before. Do you agree?
Originally posted by the lioness,:
quote:Try to find one peer review article that is not written by Clyde Winters that even mentions Dravidians and Nubian C group in the same article
Originally posted by Troll Patrol # Ish Gebor:
Yeah, thanks for the reply.
This was posted by Clyde, do you agree?
The majority of M clades in Asia are carried by Dravidian people, Dravidian people originated in Africa and belonged to the C-Group people.
quote:Now that I'm on this, anyway. What was the cranial metric like?
Originally posted by the lioness,:
quote:stop making up stuff, thanks
Originally posted by Clyde Winters:
The Mal'ta boy was Khoisan.
quote:
Originally posted by the lioness,:
http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0007447
2009
Updating Phylogeny of Mitochondrial DNA Macrohaplogroup M in India: Dispersal of Modern Human in South Asian Corridor
Adimoolam Chandrasekar et al
Published: October 13, 2009
DOI: 10.1371/journal.pone.000744
Abstract
To construct maternal phylogeny and prehistoric dispersals of modern human being in the Indian sub continent, a diverse subset of 641 complete mitochondrial DNA (mtDNA) genomes belonging to macrohaplogroup M was chosen from a total collection of 2,783 control-region sequences, sampled from 26 selected tribal populations of India. On the basis of complete mtDNA sequencing, we identified 12 new haplogroups - M53 to M64; redefined/ascertained and characterized haplogroups M2, M3, M4, M5, M6, M8′C′Z, M9, M10, M11, M12-G, D, M18, M30, M33, M35, M37, M38, M39, M40, M41, M43, M45 and M49, which were previously described by control and/or coding-region polymorphisms. Our results indicate that the mtDNA lineages reported in the present study (except East Asian lineages M8′C′Z, M9, M10, M11, M12-G, D ) are restricted to Indian region.The deep rooted lineages of macrohaplogroup ‘M’ suggest in-situ origin of these haplogroups in India. Most of these deep rooting lineages are represented by multiple ethnic/linguist groups of India. Hierarchical analysis of molecular variation (AMOVA) shows substantial subdivisions among the tribes of India (Fst = 0.16164). The current Indian mtDNA gene pool was shaped by the initial settlers and was galvanized by minor events of gene flow from the east and west to the restricted zones. Northeast Indian mtDNA pool harbors region specific lineages, other Indian lineages and East Asian lineages. We also suggest the establishment of an East Asian gene in North East India through admixture rather than replacement.
Origin of Macrohaplogroup M
L3 lineages other than M and N are absent in India and among non-African mitochondria in general [2]–[3], [49]. M, N and R haplogroups of mtDNA have no indication of an African origin. However, it is proposed that the origin of haplogroup M is in Africa [34], in view of its high frequency in Ethiopia. But in 2006, by [35] demonstrated that the presence of M1 and U6 in Africa is due to a back migration. Sequencing of 81 entire human mitochondrial DNAs belonging to haplogroups M1 and U6 revealed that these predominantly North African Clades arose in Southwestern Asia and moved together to Africa about 40,000 to 45,000 years ago. Only some sub-sets of M1a (with an estimated coalescence time of 28.8±4.9ky), U6a2 (with an estimated coalescence time of 24.0±7.3ky), and U6d (with an estimated coalescence time of 20.6±7.3ky) diffused to East and North Africa through the Levant, leaving the origin of macrohaplogroup M unresolved. Haplogroup M has been found ubiquitous in India, although its frequency is somewhat higher in southern Indian populations than in northern Indian populations and to a large extent autochthonous because neither the East nor the West Eurasian mtDNA pools include such lineages at notable frequencies [37], [58]. Our findings, (for example, deep time depth >50,000 years of western, central, southern and eastern Indian haplogroups M2, M38, M54, M58, M33, M6, M61, M62 and distribution of macrohaplogroup M) do not rule out the possibility of macrohaplogroup M arising in Indian population.
Migration routes of modern human
Recent mtDNA evidence on modern human out of Africa migration route suggests a single dispersal by a southern coastal route to India and further, to East Asia and Australia [17], [20], [22], [23], [66], [69]. The North Asian route could not get support from mtDNA due to the lack of basal M, R, N lineages in northern Asians, thereby ruling out the existence of a northern Asian route [29]–[30], [70]–[71]. Proven back migration of sub lineages of M and U into Africa [35], and the absence of L3 lineages or ancestral lineage for L3, M and N in India, leaves two issues unresolved: evidences for the southern route hypothesis from India and origin of M haplogroup. However, in the present study, the basal diversity (37 nodes) and founder ages (57,000–75,000 years) of macrohaplogroup M in India reveals initial settlement of African exodus in India. Our database also reveals evidences that Andaman islanders and Australians have ancestral maternal roots in India [24], [43].
In summary, the present study provides evidence that several Indian mtDNA M lineages are deep rooted and in situ origin. In North East India the coalescent time of East Asian lineages dates back to Last Glacial Maximum (LGM). Further, the combination of virtually all previously reported lineages from South and East Asia and our newly produced Indian complete mtDNA sequences have helped to define several novel (sub) haplogroups. The present work further ascertained previously reported haplogroups, and refined the phylogenetic tree of South Asia. This updated phylogenetic tree provides an essential reference guide for diseases, anthropological and forensic studies among Asian populations.
quote:--Adimoolam Chandrasekar et al. 2009
"The presence of M haplogroup in Ethiopia, named M1, led to the proposal that haplogroup M originated in eastern Africa, approximately 60,000 years ago, and was carried towards Asia [34].
Macrohaplogroup M is ubiquitous in India and covers more than 70 per cent of the Indian mtDNA lineages [28], [36]–[38]. Recent studies on complete mtDNA sequences (~187) tried to resolve the phylogeny of Indian macrohaplogroup M. As a result, M2, M3, M4, M5, M6 [28], [36], [39]–[40], M18, M25 [38], M30, [41], M31 [42], [24] M33, M34, M35, M36, M37, M38, M39, M40 [22], M41, M42 [43], M43 [23], [44], M45 [45], M48, M49, and M50 [46] haplogroups of M that was identified in India helped to a certain extent in understanding M genealogy in diversified Indian populations. In the above background, extensive sequencing of complete mtDNA of South Asia, particularly India, is essential for better understanding of the peopling of the non-African continents, and pathogenesis of diseases in various ethnic groups with different matrilineal backgrounds."
quote:--SUVENDU MAJI, S. KRITHIKA and T. S. VASULU (2009)
Updating Phylogeny of Mitochondrial DNA Macrohaplogroup M in India: Dispersal of Modern Human in South Asian Corridor
"Macrohaplogroup M (489-10400-14783-15043), excluding M1 which is east African, is distributed among most south, east and north Asians, Amerindians (containing a minority of north and central Amerindians and a majority of south Amerindians), and many central Asians and Melanesians."
quote:The M1 reference is your own. It is not in the papers cited. See below for recurrent mutations.
Originally posted by Troll Patrol # Ish Gebor:
Haplotypes with HVSI transitions defining 16129-16223-16249-16278-16311-16362; and 16129-16223-16234-16249-16211-16362 have been found in Thailand and among the Han Chinese (Fucharoen et al, 2001; Yao et al, 2002) and these were originally thought to be members of Haplogroup M1.
quote:The difficulty here , and one that mystifies Clyde, are "recurrent mutations." Apparently some sites are more prone to mutations than others and thus can show up in different haplotypes. Thus it is important in determining a classification that all the identifying mutated SNPs be present. These are old papers by standards of the field-- I keep emphasizing that you need to check phylotree for changes that may have happened in the interval. M1 is defined by: T195C! G6446A T6680C C12403T A12950c G16129A! T16189C! T16249C T16311C!
However, on the basis of currently available FGS sequences, carriers of these markers have been found to be in the D4a branch of Haplogroup D , the most widespread branch of M1 in East Asia (Fucharoen et al, 2001; Yao et al, 2002). The transitions 16129,16189,16249 and 16311 are known to be recurrent in various branches of Haplogroup M, especially M1 and D4.]
quote:Fucharoen, et al .2001 “Mitochondrial DNA polymorphisms in Thailand,” J Hum Genet 46:115–125
p 638 Among the three R* haplotypes that could not be classified as B or R9, two bear a mutation motif of 185-189-10398-16189-16311, similar to the motif of B5, but were found to lack the 9-bp deletion.
p. 641 Table 1 D4a 129 223 249 278 311 362
p 645 Type 1 (16223-[16311-16357) matches haplotypes from M10 (one sampled in Liaoning and another one in Yunnan).
p. 646 In contrast, sequence 19 (16223, 146-263) has no close companion (at distance two or fewer mutational steps) in the Han
The haplogroup affiliations of the resulting nine haplotypes, except for type 9 (16256-16278- 16295), can be recognized by following our classification strategy. Type 1 (16223-16311-16357) matches haplotypes from M10 (one sampled in Liaoning and another one in Yunnan), and type 7 (16284) matches a B4b haplotype from Liaoning. The other six types have one-step neighbors in the Han mtDNA database: type 2 (16223-16234-16290-16319) is thus related to A haplotypes from Wuhan and Yunnan; type 3 (16223-16298-16319- 16355) to M8a haplotypes from Qingdao and Wuhan; type 4 (16223-16266-16274-16362) to a D4 haplotype from Liaoning and to D5a haplotypes from Liaoning, Wuhan, Xinjiang, and Qingdao; type 6 (16223-16278- 16362) to two G2 haplotypes and type 8 (16223-16245- 16362-16368) to one D4 haplotype, all from Liaoning; finally, type 5 (16223-16357) is a one-step descendant of the matched M10 type 1 (but, alternatively, it would also be a one-step neighbor of an M* haplotype from Qingdao)
quote:As you can see in neither paper reports all the M1 identifiers.
p. 121 A G-to-A transition at 16129 occurred in the majority of lineages from C2 (71%) and C6(88%), whereas these two clusters appeared at discrete positions in the phylogenetic tree. Thus, the above three polymorphisms are due to either recurrent mutations or ancient polymorphisms.
Table 2
Cluster
C2 52 16108:C/T (40%) 16129:G/A (71%) 16162:A/G (44%) 16172:T/C (63%) 16304:T/C (92%) 16519:T/C (88%)
C5 10 16223:C/T (100%) 16278:C/T (60%) 16519:T/C (40%)
A C-to-T transition at 16223 (nucleotide position in the reference sequence of Anderson et al. 1981) was shared by most members in clusters C3a and C4 through C8, while this polymorphism was virtually absent in clusters C1, C2, and C3b.
quote:There are NO African genes in Thailand or China.
As you can see geneticist change the name of haplogroups to create confusion and deny relationships between Africans and non-Africans.
Another example is V88, although it is R1-M173, it was given another name to make it appear to be different from the R1 group.
In summary CHINESE carry African clades.
quote:There is no such thing as recurrent mutations. I proved this point in my articles on M1 mutations in India. See: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3168144/
Originally posted by Quetzalcoatl:
The difficulty here , and one that mystifies Clyde, are "recurrent mutations." Apparently some sites are more prone to mutations than others and thus can show up in different haplotypes. Thus it is important in determining a classification that all the identifying mutated SNPs be present. These are old papers by standards of the field-- I keep emphasizing that you need to check phylotree for changes that may have happened in the interval. M1 is defined by: T195C! G6446A T6680C C12403T A12950c G16129A! T16189C! T16249C T16311C! . [/QB]
quote:Again delusions of grandeur. A google search for " DNA recurrent mutations" turns up 12,800,000 results. If you really had overturned the existence of recurrent mutations with your cherry-picked literature review, you should be getting a Nobel Prize. I'm sure that the American Journal of Human Evolution would be glad to publish such an earth shattering piece of research.
Originally posted by Clyde Winters:
quote:There is no such thing as recurrent mutations. I proved this point in my articles on M1 mutations in India. See: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3168144/
Originally posted by Quetzalcoatl:
The difficulty here , and one that mystifies Clyde, are "recurrent mutations." Apparently some sites are more prone to mutations than others and thus can show up in different haplotypes. Thus it is important in determining a classification that all the identifying mutated SNPs be present. These are old papers by standards of the field-- I keep emphasizing that you need to check phylotree for changes that may have happened in the interval. M1 is defined by: T195C! G6446A T6680C C12403T A12950c G16129A! T16189C! T16249C T16311C! .
LOL. This article was peer reviewed and appeared in a publication recognized by NCBI.
Also see: Advantageous Alleles, Parallel Adaptation, Geographic Location and Sickle Cell Anemia among Africans and Indians http://www.academia.edu/3036807/Advantageous_Alleles_Parallel_Adaptation_Geographic_Location_and_Sickle_Cell_Anemia_among_Africans_and_Indians
. [/QB]
quote:How can it be my own reference, when I cited Clyde Winters on this and asked you your opinion on it. While you responded with you other troll account.."
Originally posted by Quetzalcoatl:
The M1 reference is your own. It is not in the papers cited. See below for recurrent mutations.
quote:I agree, these are old papers.
Originally posted by Quetzalcoatl:
The difficulty here , and one that mystifies Clyde, are "recurrent
mutations." Apparently some sites are more prone to mutations than others and thus can show up in different haplotypes. Thus it is important in determining a classification that all the identifying mutated SNPs be present. These are old papers by standards of the field--
I keep emphasizing that you need to check phylotree for changes that may have happened in the interval. M1 is defined by: T195C! G6446A T6680C C12403T A12950c G16129A! T16189C! T16249C T16311C!
Yao, Y-G, et al 2002 “Phylogeographic Differentiation of Mitochondrial DNA in Han Chinese,” Am. J. Human Genetics 70:635-651
polymorphisms 16129 and 16249 were not found
quote:http://www.cagetti.com/Genetics/reportmtdna.pdf
Phylogenetic Tree of Global Mitochondrial DNA
Every person alive today can trace their maternal lineage to a single woman who lived in Africa approximately 160,000 years ago. She has been called the "Mitochondrial Eve" - there is no relation at all to the biblical Eve. As our ancestors migrated out of Africa and settled in Europe, Asia and the Americas, mutations occurred that became part of the genetic make-up of particular geographical populations. This diagram depicts how these mutations led to branching in the phylogenetic tree of mtDNA. The letters in boxes represent mitochondrial haplogroups, or "clans", that are comprised of people with similar lineages. The numbers on the lines are positions within the mitochondrial genome at which polymorphisms (mutations) are found that define the haplogroup.
The numbers next to some of the boxes are approximate coalescent times, i.e., the time in the past that the haplogroup originated. Coalescent times are in thousands of years (Kivisild, 2006).
quote:--Kivisild Investigative Genetics (2015) 6:3 DOI 10.1186/s13323-015-0022-2
Distribution of variation in mtDNA genomes among human populations
Compared to the estimates based on autosomal data the observed differences in mitochondrial sequences among human populations on a global scale are significantly higher and second only to the differences based on Y chromosomes, with Africa showing the highest within region diversity and Native Americans having the lowest [56]. As it has been repeatedly shown with ever increas- ing sample sizes that are reaching tens of thousands of individuals now [68], the root of the mtDNA phylogeny and the most diverse branches are restricted to African populations (Figure 2). Using the maximum molecular resolution enabled by the analysis of whole mtDNA ge- nomes, the first seven bifurcations in this tree, in fact, define the distinction of strictly sub-Saharan African branches (L0-L6) from those that are shared by Africans and non-African populations. Analyses of whole mtDNA sequences of sub-Saharan Africans have revealed early, ca 90 to 150 thousand years (ky) old divergence of the L0d and L0k lineages that are specific to the Khoisan populations from South Africa and it has been estimated that during this time period at least six additional line- ages existed in Africa with living descendants [53,54]. In contrast to the overall high basal clade diversity and geographic structure some terminal branches from haplogroups L0a, L1c, L2a, and L3e show recent co- alescent times and wide geographical distribution in Africa, likely due to the recent Bantu expansion [70-72].
[...]
The fact that virtually every non-African mtDNA lineage derives from just one of the two sub-clades of the African haplogroup L3 (Figure 2) has been inter- preted as an evidence of a major bottleneck of mtDNA diversity at the onset of the out of Africa dispersal [74]. The magnitude of this bottleneck has been estimated from the whole mtDNA sequence data yielding the esti- mates of the effective population size which range be- tween several hundred [75] and only few tens of females [56]. The separation of these two sub-clades, M and N, from their African sister-clades in L3 can be dated back to 62 to 95 kya [48] whereas the internal coalescent time estimates of the M and N founders have been estimated in the range of 40 to 70 ky [26,28,75] and suggest that their dispersal occurred probably after rather than before the eruption of Mount Toba 74 kya in Indonesia, one of the Earth’s largest known volcanic events in human his- tory. Archaeological evidence from Jurreru River valley, India, has shown the presence of artefacts right above and below the layers of ash associated with the Toba eruption [76]. It is not clear whether the makers of these artefacts were archaic or anatomically modern humans. As in case of the global TMRCA estimate considered above the wide error ranges around the age estimates of haplogroups M and N reflect primarily the uncertainties of the mutation rate - in relative terms, the age estimates of M and N, as determined from whole mtDNA se- quences form approximately one third of the total depth of the global mtDNA tree. Claims for relatively recent, post-Toba, time depth of the non-African founder- haplogroups have been recently supported by the aDNA evidence of the 45 kya Ust-Ishim skeleton whose whole mtDNA sequence falls at the root of haplogroup R [50]. While haplogroups M and N are widely spread in Asia, Australia, Oceania and Americas, the geographic distri- bution of each of their sub-clades has more specific re- gional configuration (Figure 2).
quote:I had to look it up, ...
Originally posted by the lioness,:
quote:Try to find one peer review article that is not written by Clyde Winters that even mentions Dravidians and Nubian C group in the same article
Originally posted by Troll Patrol # Ish Gebor:
Yeah, thanks for the reply.
This was posted by Clyde, do you agree?
The majority of M clades in Asia are carried by Dravidian people, Dravidian people originated in Africa and belonged to the C-Group people.
quote:As demonstrated by Troll Patrol # Ish Gebor posting on Dravidian language being an African language, and my own postings on the relationship between skin color and vitamin D absorption (None).
Originally posted by Quetzalcoatl:
If you really had overturned the existence of recurrent mutations with your cherry-picked literature review, you should be getting a Nobel Prize. I'm sure that the American Journal of Human Evolution would be glad to publish such an earth shattering piece of research.
quote:One is written by an Lecturer at the University College of Art and Design, (M.A.) The Islamia University Bahawalpur in Pakistan.
Originally posted by Troll Patrol # Ish Gebor:
quote:I had to look it up, ...
Originally posted by the lioness,:
quote:Try to find one peer review article that is not written by Clyde Winters that even mentions Dravidians and Nubian C group in the same article
Originally posted by Troll Patrol # Ish Gebor:
Yeah, thanks for the reply.
This was posted by Clyde, do you agree?
The majority of M clades in Asia are carried by Dravidian people, Dravidian people originated in Africa and belonged to the C-Group people.
Dravidian and Nubian
http://www.jstor.org/stable/593167?seq=1#page_scan_tab_contents
IOSR Journal Of Humanities And Social Science (IOSR-JHSS) Volume 20, Issue 2, Ver. V (Feb. 2015), PP 66-72
e-ISSN: 2279-0837, p-ISSN: 2279-0845. www.iosrjournals.org
Comparative Visual Analysis of Symbolic and Illegible Indus Valley Script with Other Languages
Mrs. Maria Ansari1, Mr. Farjad Faiz2, Ms. Amna Ansari3
1,2, University College of Art and Design, The Islamia University Bahawalpur, Pakistan 3.College of Art and Design, Punjab University Lahore, Pakistan
http://iosrjournals.org/iosr-jhss/papers/Vol20-issue2/Version-5/K020256672.pdf
Dravidian costumes and household articles
Dr.A.Sagayadoss,
Director Council of Agri Geo Environmental Research Bengaluru
http://www.academiaandsocietyjournal.com/uploads/3/8/8/5/38850075/1.__dravidian_costumes_and_household_articles.pdf
Apparently there is support for Clyde's theory.
quote:You are right the Dravidians traveled to number of locations in Eurasia after they left the Proto-Saharan along with the Mande speaking people.You can find out more about their travels and settlements in the articles below and my book on the Tamil in Central Asia. Enjoy
Originally posted by the lioness,:
quote:One is written by an Lecturer at the University College of Art and Design, (M.A.) The Islamia University Bahawalpur in Pakistan.
Originally posted by Troll Patrol # Ish Gebor:
quote:I had to look it up, ...
Originally posted by the lioness,:
quote:Try to find one peer review article that is not written by Clyde Winters that even mentions Dravidians and Nubian C group in the same article
Originally posted by Troll Patrol # Ish Gebor:
Yeah, thanks for the reply.
This was posted by Clyde, do you agree?
The majority of M clades in Asia are carried by Dravidian people, Dravidian people originated in Africa and belonged to the C-Group people.
Dravidian and Nubian
http://www.jstor.org/stable/593167?seq=1#page_scan_tab_contents
IOSR Journal Of Humanities And Social Science (IOSR-JHSS) Volume 20, Issue 2, Ver. V (Feb. 2015), PP 66-72
e-ISSN: 2279-0837, p-ISSN: 2279-0845. www.iosrjournals.org
Comparative Visual Analysis of Symbolic and Illegible Indus Valley Script with Other Languages
Mrs. Maria Ansari1, Mr. Farjad Faiz2, Ms. Amna Ansari3
1,2, University College of Art and Design, The Islamia University Bahawalpur, Pakistan 3.College of Art and Design, Punjab University Lahore, Pakistan
http://iosrjournals.org/iosr-jhss/papers/Vol20-issue2/Version-5/K020256672.pdf
Dravidian costumes and household articles
Dr.A.Sagayadoss,
Director Council of Agri Geo Environmental Research Bengaluru
http://www.academiaandsocietyjournal.com/uploads/3/8/8/5/38850075/1.__dravidian_costumes_and_household_articles.pdf
Apparently there is support for Clyde's theory.
She mentions Clyde's "deciperment"
The other is a 1932 article , I haven't read called
"Dravidian and Nubian" by an Edwin H Tuttle
he also wrote
" Finnic and Dravidian" in 1911 and
"Turkish and Dravidian" in 1912
Appaently these Dravidians were all over the place


quote:Sorry. Sometimes when the posts say "quote" too many times it is not clear to whom you are responding.my answer was meant for Clyde.
Originally posted by Troll Patrol # Ish Gebor:
quote:How can it be my own reference, when I cited Clyde Winters on this and asked you your opinion on it. While you responded with you other troll account.."
Originally posted by Quetzalcoatl:
The M1 reference is your own. It is not in the papers cited. See below for recurrent mutations.
I looked up the references myself:
--Fucharoen, et al .
“Mitochondrial DNA polymorphisms in Thailand,” J Hum Genet 46:115–125
http://download.bioon.com.cn/upload/20110610/2011061071.pdf
-- Yao et a.
Phylogeographic Differentiation of Mitochondrial DNA in Han Chinese
Am J Hum Genet. 2002 Mar; 70(3): 635–651.
Published online 2002 Feb 8. doi: 10.1086/338999
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC384943/
quote:I agree, these are old papers.
Originally posted by Quetzalcoatl:
The difficulty here , and one that mystifies Clyde, are "recurrent
mutations." Apparently some sites are more prone to mutations than others and thus can show up in different haplotypes. Thus it is important in determining a classification that all the identifying mutated SNPs be present. These are old papers by standards of the field--
I keep emphasizing that you need to check phylotree for changes that may have happened in the interval. M1 is defined by: T195C! G6446A T6680C C12403T A12950c G16129A! T16189C! T16249C T16311C!
Yao, Y-G, et al 2002 “Phylogeographic Differentiation of Mitochondrial DNA in Han Chinese,” Am. J. Human Genetics 70:635-651
polymorphisms 16129 and 16249 were not found
So these "recurrent mutations" (16129, 16223 and 16311) have never been found in Africa?
mtDNA Haplogroup Specific Control Region Mutation Motifs
http://mtmanager.yonsei.ac.kr/help/MutationMotifs.pdf
C16223T
T16311C
http://www.ncbi.nlm.nih.gov/pubmed/21042748
Apparently Clyde has more papers out, which have been recognized by NCBI.
http://www.ncbi.nlm.nih.gov/pubmed/?term=Winters%20C%5Bauth%5D
quote:No. My original question: "Try to find one peer review article that is not written by Clyde Winters that even mentions Dravidians and Nubian C group in the same article."
Originally posted by Troll Patrol # Ish Gebor:
quote:I had to look it up, ...
Originally posted by the lioness,:
quote:Try to find one peer review article that is not written by Clyde Winters that even mentions Dravidians and Nubian C group in the same article
Originally posted by Troll Patrol # Ish Gebor:
Yeah, thanks for the reply.
This was posted by Clyde, do you agree?
The majority of M clades in Asia are carried by Dravidian people, Dravidian people originated in Africa and belonged to the C-Group people.
Dravidian and Nubian
http://www.jstor.org/stable/593167?seq=1#page_scan_tab_contents
IOSR Journal Of Humanities And Social Science (IOSR-JHSS) Volume 20, Issue 2, Ver. V (Feb. 2015), PP 66-72
e-ISSN: 2279-0837, p-ISSN: 2279-0845. www.iosrjournals.org
Comparative Visual Analysis of Symbolic and Illegible Indus Valley Script with Other Languages
Mrs. Maria Ansari1, Mr. Farjad Faiz2, Ms. Amna Ansari3
1,2, University College of Art and Design, The Islamia University Bahawalpur, Pakistan 3.College of Art and Design, Punjab University Lahore, Pakistan
http://iosrjournals.org/iosr-jhss/papers/Vol20-issue2/Version-5/K020256672.pdf
Dravidian costumes and household articles
Dr.A.Sagayadoss,
Director Council of Agri Geo Environmental Research Bengaluru
http://www.academiaandsocietyjournal.com/uploads/3/8/8/5/38850075/1.__dravidian_costumes_and_household_articles.pdf
Apparently there is support for Clyde's theory.
quote:Which part of Albino opinion/acceptance/rating, means nothing in the absence of rational fact based refutation, don't you understand? Prove your points with facts, not Albino opinions. Otherwise, go peddle your sh1t to Albinos and Negroes.
Originally posted by Quetzalcoatl:
The only journal I found in which Winters published original articles (not comments or letters which as I have shown are not reviewed) was the International Journal of Human genetics. This journal asked the author to send in the names of 3 proposed reviewers (in most peer reviewed journals editors choose the reviewers). Its impact factor is.0155
Journal impact factor
Int. J Hum. genetics 0.155
PlOs ONE 4.411
BSC genetics 2.439
BSC genomics 3.716
Am. J Human genetics 10.987
Science 31.027
Nature 42.351
quote:1) You asked to look up peer reviewed sources, which mentioned Dravidians and Nubian (C) in the same article, which I did.
Originally posted by the lioness,:
quote:One is written by an Lecturer at the University College of Art and Design, (M.A.) The Islamia University Bahawalpur in Pakistan.
Originally posted by Troll Patrol # Ish Gebor:
quote:I had to look it up, ...
Originally posted by the lioness,:
quote:Try to find one peer review article that is not written by Clyde Winters that even mentions Dravidians and Nubian C group in the same article
Originally posted by Troll Patrol # Ish Gebor:
Yeah, thanks for the reply.
This was posted by Clyde, do you agree?
The majority of M clades in Asia are carried by Dravidian people, Dravidian people originated in Africa and belonged to the C-Group people.
Dravidian and Nubian
http://www.jstor.org/stable/593167?seq=1#page_scan_tab_contents
IOSR Journal Of Humanities And Social Science (IOSR-JHSS) Volume 20, Issue 2, Ver. V (Feb. 2015), PP 66-72
e-ISSN: 2279-0837, p-ISSN: 2279-0845. www.iosrjournals.org
Comparative Visual Analysis of Symbolic and Illegible Indus Valley Script with Other Languages
Mrs. Maria Ansari1, Mr. Farjad Faiz2, Ms. Amna Ansari3
1,2, University College of Art and Design, The Islamia University Bahawalpur, Pakistan 3.College of Art and Design, Punjab University Lahore, Pakistan
http://iosrjournals.org/iosr-jhss/papers/Vol20-issue2/Version-5/K020256672.pdf
Dravidian costumes and household articles
Dr.A.Sagayadoss,
Director Council of Agri Geo Environmental Research Bengaluru
http://www.academiaandsocietyjournal.com/uploads/3/8/8/5/38850075/1.__dravidian_costumes_and_household_articles.pdf
Apparently there is support for Clyde's theory.
She mentions Clyde's "deciperment"
Another article is by
Dr.A.Sagayadoss,Independent Researcher with a Ph.D in Industrial waste water treatments
The other is a 1932 article , I haven't read called
"Dravidian and Nubian" by an Edwin H Tuttle
he also wrote
" Finnic and Dravidian" in 1911 and
"Turkish and Dravidian" in 1912
Appaently these Dravidians were all over the place
Clyde's gone mainstream
quote:http://es.about.jstor.org/jstor-help-support/395974/395978/395999/
Is all content on JSTOR peer reviewed?
While nearly all of the journals collected in JSTOR are peer-reviewed publications, our archives do contain some specific primary materials (like some journals in the Ireland Collection and the 19th Century British Pamphlet Collection). Also, some journal content is much older than today's standard peer-review process. This means that, though all the information in JSTOR is held to a scholarly standard, not all of the publications are technically "peer-reviewed." At the current time there is no way to search JSTOR for only peer-reviewed publications. We often find that if you have questions concerning the academic legitimacy of a particular journal or book, your institution's librarian or your course instructor may be best able to answer those inquiries.
quote:http://iosrjournals.org/iosr-jhss.html
IOSR Journal of Humanities and Social Science is a double blind peer reviewed International Journal edited by International Organization of Scientific Research (IOSR).The Journal provides a common forum where all aspects of humanities and social sciences are presented. IOSR-JHSS publishes original papers, review papers, conceptual framework, analytical and simulation models, case studies, empirical research, technical notes etc.
quote:http://www.dialoguesociety.org/publications/academia/829-journal-of-dialogue-studies.html
The Journal of Dialogue Studies is a multidisciplinary, peer-reviewed academic journal published twice a year.
quote:It becomes confusing when you reply under both accounts. Quetzalcoatl and your troll account.
Originally posted by Quetzalcoatl:
My original question: "Try to find one peer review article that is not written by Clyde Winters that even mentions Dravidians and Nubian C group in the same article."
The second paper is an open (not reviewed ) access paper. The last paper is uploaded by the author to Academia.edu, and not reviewed or published in a journal.
The first article 1932 was published before Clyde was born. I did not want to spend money to see if the C-group was the Nubian connection.
quote:How can this be confusing. When I clearly addressed the question towards you.
Originally posted by Quetzalcoatl:
quote:Sorry. Sometimes when the posts say "quote" too many times it is not clear to whom you are responding.my answer was meant for Clyde.
Originally posted by Troll Patrol # Ish Gebor:
quote:How can it be my own reference, when I cited Clyde Winters on this and asked you your opinion on it. While you responded with you other troll account.."
Originally posted by Quetzalcoatl:
The M1 reference is your own. It is not in the papers cited. See below for recurrent mutations.
I looked up the references myself:
--Fucharoen, et al .
“Mitochondrial DNA polymorphisms in Thailand,” J Hum Genet 46:115–125
http://download.bioon.com.cn/upload/20110610/2011061071.pdf
-- Yao et a.
Phylogeographic Differentiation of Mitochondrial DNA in Han Chinese
Am J Hum Genet. 2002 Mar; 70(3): 635–651.
Published online 2002 Feb 8. doi: 10.1086/338999
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC384943/
quote:I agree, these are old papers.
Originally posted by Quetzalcoatl:
The difficulty here , and one that mystifies Clyde, are "recurrent
mutations." Apparently some sites are more prone to mutations than others and thus can show up in different haplotypes. Thus it is important in determining a classification that all the identifying mutated SNPs be present. These are old papers by standards of the field--
I keep emphasizing that you need to check phylotree for changes that may have happened in the interval. M1 is defined by: T195C! G6446A T6680C C12403T A12950c G16129A! T16189C! T16249C T16311C!
Yao, Y-G, et al 2002 “Phylogeographic Differentiation of Mitochondrial DNA in Han Chinese,” Am. J. Human Genetics 70:635-651
polymorphisms 16129 and 16249 were not found
So these "recurrent mutations" (16129, 16223 and 16311) have never been found in Africa?
mtDNA Haplogroup Specific Control Region Mutation Motifs
http://mtmanager.yonsei.ac.kr/help/MutationMotifs.pdf
C16223T
T16311C
http://www.ncbi.nlm.nih.gov/pubmed/21042748
Apparently Clyde has more papers out, which have been recognized by NCBI.
http://www.ncbi.nlm.nih.gov/pubmed/?term=Winters%20C%5Bauth%5D
Of course 16129, 16223, 163211 are found in Africa! They are the defining SNP's for M1 an African haplotype.
Being listed in the NCI is not any kind of official recognition. If one is interested in what the quality of a particular journal, the place to look is the listing of impact factor of journals
http://www.citefactor.org/journal-impact-factor-list-2014_I.html
The only journal I found in which Winters published original articles (not comments or letters which as I have shown are not reviewed) was the International Journal of Human genetics. This journal asked the author to send in the names of 3 proposed reviewers (in most peer reviewed journals editors choose the reviewers). Its impact factor is.0155
Journal impact factor
Int. J Hum. genetics 0.155
PlOs ONE 4.411
BSC genetics 2.439
BSC genomics 3.716
Am. J Human genetics 10.987
Science 31.027
Nature 42.351
quote:--Adimoolam Chandrasekar et al. 2009
"The presence of M haplogroup in Ethiopia, named M1, led to the proposal that haplogroup M originated in eastern Africa, approximately 60,000 years ago, and was carried towards Asia [34].
Macrohaplogroup M is ubiquitous in India and covers more than 70 per cent of the Indian mtDNA lineages [28], [36]–[38]. Recent studies on complete mtDNA sequences (~187) tried to resolve the phylogeny of Indian macrohaplogroup M. As a result, M2, M3, M4, M5, M6 [28], [36], [39]–[40], M18, M25 [38], M30, [41], M31 [42], [24] M33, M34, M35, M36, M37, M38, M39, M40 [22], M41, M42 [43], M43 [23], [44], M45 [45], M48, M49, and M50 [46] haplogroups of M that was identified in India helped to a certain extent in understanding M genealogy in diversified Indian populations. In the above background, extensive sequencing of complete mtDNA of South Asia, particularly India, is essential for better understanding of the peopling of the non-African continents, and pathogenesis of diseases in various ethnic groups with different matrilineal backgrounds."
quote:What in the world are you talking about? What troll account?
Originally posted by Troll Patrol # Ish Gebor:
[]It becomes confusing when you reply under both accounts. Quetzalcoatl and your troll account.
quote:]
[Winters]
posted 21 February, 2008 03:07 PM
[QUOTE]quote:
Originally posted by Clyde Winters:
mtDNA research supports an African influence in Near Oceania. For example researchers have found that the Tanzanian M1 haplogroup cluster with people from Oceania (Gonder et al, 2006).
In addition, the M1 mutations 16129,16189,16249 and 16311 are found in many southeast and East Asian haplogroups (Fucharoen et al, 2001; Yao et al, 2002).
This molecular evidence further supports the Neolithic skeletal evidence of a recent migration of Africans to the Pacific and east Asian region after the initial exit from Africa of AMH.
quote:[Rasol]
Finally, our limited genetic data from Tanzanians belonging to haplogroups M1, N1, and J suggest 2 alternatives that are not mutually exclusive. Populations in Tanzania may have been important in the migration of modern humans from Africa to other regions, as noted in previous studies of other populations in eastern Africa (Quintana- Murci et al. 1999). For example, mtDNAs of Tanzanians belonging to haplogroup M1 cluster with peoples from Oceania, whereas Tanzanian mtDNAs belonging to haplogroup N1 and J cluster with peoples of Middle Eastern and Eurasian origin. However, the presence of haplogroups N1 and J in Tanzania suggest ‘‘back’’ migration from the Middle East or Eurasia into eastern Africa, which has been inferred from previous studies of other populations in eastern Africa (Kivisild et al. 2004). These results are intriguing and suggest that the role of Tanzanians in the migration of modern humans within and out of Africa should be analyzed in greater detail after more extensive data collection, particularly from analysis of Y-, X-, and autosomal chromosome markers. Our analyses of African mtDNAs suggest populations in eastern Africa have played an important and persistent role in the origin and diversification of modern humans.
quote:[Ortiz de Montellano]
Meaning: Tanzana has M1, N1 and J haplotypes. M, *NOT* M1 is found in Oceana. N and J [including but not limited to N1] is found South West Asia and Europe.
Nowhere is anything said about M1 found in Oceana.
quote:.
Not only is Rasol correct, but Gonder et al, as you can see from the title of the paper, " “Whole mtDNA Genome Sequence Analysis of Ancient African Lineages" is not dealing with recent gene flow from Africa but rather with the initial Out of Africa expansion of mode humans
quote:subsequently I wrote to Dr. Gonder to verify my interpretation of her paper and Winters’s error
mtDNAs belonging to haplogroups M and N form 2 monophyletic clades (fig. 2A). These 2 M and N haplogroup clades included a few Tanzanians (belonging to haplogroups M1, M, N1, and J), suggesting possible recent gene flow back into Africa and/or that ancestors of the Tanzanian populations may have been a source of migration of modern humans from Africa to other regions (fig. 2B). Or from Gonder's phylogenetic tree, the date of the last common African and non-African haplotypes
The age of the youngest node containing both African and non-African sequences (node S) is 94.3 6 9.9 kya and represents an upper bound time estimate for an exodus out of Africa. isolated quotes are meaningless unless they are set in context with the entire paper.
quote:Winters is referring to the following passage in your paper:
Date: Mon, 17 Mar 2008 09:34:07 -0400
Subject: Re: supplementary material
From: Katy Gonder mgonder@umd.edu
To: Bernard Ortiz de Montellano <bortiz@earthlink.net>
Thread-Topic: supplementary material
Hi,
You are correct, we were referring to the initial migration of out of Africa. Cluster together....I haven’t looked at the tree in detail for some time, but as I remember, the TZ M formed part of the “basal” lineages containing both African and non-African mtDNA genomes. Many of the non-African genomes were of oceanic origin. [/b] We were definitely not referring to anything that happened with the last few thousand years.[/b]
Hope that helps.
Katy
On 3/16/08 11:20 PM, "Bernard Ortiz de Montellano" <bortiz@earthlink.net> wrote:
Hi
Thank you so much for sending me the supplementary information for your paper. I have no idea why Molecular Evolution and Biology made it impossible to download. Unfortunately, they do not answer my basic question. Clyde Winters has been quoting your paper in support of his theory that the Mande migrated out of Africa a few thousand years ago ,i.e. in - BioEssays 29:497-498, 2007, Winters writes that your paper supports his claims that Mande speakers migrated from Africa to become the Dravidians of India. Quote: "Anna Oliviera et al. argue that M1 must have originated in West Asia, because none of the Asian M haplogroups harbor any distinguishing East African root mutations. (30) They claim that the presence of any East African M1 root mutations in Asian-specific clades suggest a recent arrival of M1; and that the absence of M1 root mutations among Eurasian sister clades indicate a back migration into East Africa of HG M1. (30)”
[Winters] Oliviera et al. claim that East African M1 root mutations are absent in Eurasian M sister clades is not supported by the evidence. (36) For example, Gondar [sic] et al. make it clear that the Tanzanian M1 haplogroup cluster with people from Oceania. In addition, Roychoudhury et al. noted nucleolides shared by East African M1, and Indian M haplogroups include HG M4 at 16311; HG M5 at 16,129; and HG M34 at 16,249."
quote:[Ortiz de Montellano]
"Populations in Tanzania may have been important in the migration of modern humans from Africa to other regions, as noted in previous studies of other populations in eastern Africa (Quintana- Murci et al. 1999). For example, mtDNAs of Tanzanians belonging to haplogroup M1 cluster with peoples from Oceania, whereas Tanzanian mtDNAs belonging to haplogroup N1 and J cluster with peoples of Middle Eastern and Eurasian origin. However, the presence of haplogroups N1 and J in Tanzania suggest ''back'' migration from the Middle East or Eurasia into eastern Africa, which has been inferred from previous studies of other populations in eastern Africa (Kivisild et al. 2004). These results are intriguing and suggest that the role of Tanzanians in the migration of modern humans within and out of Africa should be analyzed in greater detail after more extensive data collection, particularly from analysis of Y-, X-, and autosomal chromosome markers. Our analyses of African mtDNAs suggest populations in eastern Africa have played an important and persistent role in the origin and diversification of modern humans.
quote:I wrote:
"Populations in Tanzania may have been important in the migration of modern humans from Africa to other regions, as noted in previous studies of other populations in eastern Africa (Quintana- Murci et al. 1999). For example, mtDNAs of Tanzanians belonging to haplogroup M1 cluster with peoples from Oceania, whereas Tanzanian mtDNAs belonging to haplogroup N1 and J cluster with peoples of Middle Eastern and Eurasian origin. However, the presence of haplogroups N1 and J in Tanzania suggest ''back'' migration from the Middle East or Eurasia into eastern Africa, which has been inferred from previous studies of other populations in eastern Africa (Kivisild et al. 2004). These results are intriguing and suggest that the role of Tanzanians in the migration of modern humans within and out of Africa should be analyzed in greater detail after more extensive data collection, particularly from analysis of Y-, X-, and autosomal chromosome markers. Our analyses of African mtDNAs suggest populations in eastern Africa have played an important and persistent role in the origin and diversification of modern humans.
quote:As you can see I did not misquote anything. I repeated her claim that "Tanzanians belonging to haplogroup M1 cluster with peoples from Oceania".
mtDNA research supports an African influence in Near Oceania. For example researchers have found that the Tanzanian M1 haplogroup cluster with people from Oceania (Gonder et al, 2006).
quote:That other account, you responded with.
Originally posted by Quetzalcoatl:
quote:What in the world are you talking about? What troll account?
Originally posted by Troll Patrol # Ish Gebor:
[]It becomes confusing when you reply under both accounts. Quetzalcoatl and your troll account.
quote:I am not the Lioness.
Originally posted by Ish Gebor:
quote:That other account, you responded with.
Originally posted by Quetzalcoatl:
quote:What in the world are you talking about? What troll account?
Originally posted by Troll Patrol # Ish Gebor:
[]It becomes confusing when you reply under both accounts. Quetzalcoatl and your troll account.
http://www.egyptsearch.com/forums/ultimatebb.cgi?ubb=get_topic;f=15;t=010625;p=4#000174
quote:The name is Gonder not Gondor.
Originally posted by Clyde Winters:
Bernard Montellano you're such a liar and fake . Gondor wrote
quote:I wrote:
"Populations in Tanzania may have been important in the migration of modern humans from Africa to other regions, as noted in previous studies of other populations in eastern Africa (Quintana- Murci et al. 1999). For example, mtDNAs of Tanzanians belonging to haplogroup M1 cluster with peoples from Oceania, whereas Tanzanian mtDNAs belonging to haplogroup N1 and J cluster with peoples of Middle Eastern and Eurasian origin. However, the presence of haplogroups N1 and J in Tanzania suggest ''back'' migration from the Middle East or Eurasia into eastern Africa, which has been inferred from previous studies of other populations in eastern Africa (Kivisild et al. 2004). These results are intriguing and suggest that the role of Tanzanians in the migration of modern humans within and out of Africa should be analyzed in greater detail after more extensive data collection, particularly from analysis of Y-, X-, and autosomal chromosome markers. Our analyses of African mtDNAs suggest populations in eastern Africa have played an important and persistent role in the origin and diversification of modern humans.
quote:As you can see I did not misquote anything. I repeated her claim that "Tanzanians belonging to haplogroup M1 cluster with peoples from Oceania".
mtDNA research supports an African influence in Near Oceania. For example researchers have found that the Tanzanian M1 haplogroup cluster with people from Oceania (Gonder et al, 2006).
You are too stupid. The paper was not about ancient mtDNA. Gondor was reporting on the M1 found in modern Tanzanians and Oceanians you ignorant moron.
Montellano, you are a liar and Great Deceiver.
.
quote:Here you are using Gonder’s paper as if she supported your claim—which she does not. This is academic malfeasance, and it would never get by a real peer review.
The Dravidians and Manding people originated in Africa. They belonged to the C-Group culture of Nubia (Winters, 2007). After 3000 BC they began to migrate out of Africa into Eurasia (Winters, 2008). The languages of Dravidian and African speakers are genetically related (Winters, 2007, 2008). The Dravidians and Africans also share genes (Winters, 2008b, 2010, 2010b).
. . . . . .
The transitions 16129, 16189, 16249 and 16311 are known to be recurrent in various branches of Haplogroup M, especially M1 and D4. Gonder et al., (2006) for example, noted that the mtDNAs of Tanzanians belonging to haplogroup M1 cluster with peoples from Oceania.
quote:Friedlander also objected to your comments on his paper:
The intimate relations between Proto-Polynesians and Proto-Melanesians back to Shang times in China (Chang, 1964; Winters 1985, 1986), supports the hypothesis of an ancient spread of Melanesian mtDNA among (Proto-) Polynesians. Some researchers have assumed that this genetic sharing took place in Fiji (Fredlaender, 2007), but it is more probable that the sharing of Y chromosome and mtDNA types took place in mainland East Asia. This would explain the differences between shared Asian and Melanesian haplotypes and haplogroups in the Pacific.
quote:
We did find unequivocal genetic evidence of ties between Polynesians, Micronesians, and Taiwan Aborigines (as well as East Asians). Papuan-speaking groups in Island Melanesia are very distinctive, but do not seem to be especially related to Africans in spite of some similarities in appearances.
Regarding the craniometric evidence, Michael Pietrusewsky's work in Asia and the Pacific is the authoritative contemporary reference, although I believe his interpretation of Island Melanesian "intermediate" status should be altered. However, I am certain he would reject any argument that there is any evidence of recent African influence in East Asia, much less Island Melanesia.]
quote:It came off as such.
Originally posted by Quetzalcoatl:
quote:I am not the Lioness.
Originally posted by Ish Gebor:
quote:That other account, you responded with.
Originally posted by Quetzalcoatl:
quote:What in the world are you talking about? What troll account?
Originally posted by Troll Patrol # Ish Gebor:
[]It becomes confusing when you reply under both accounts. Quetzalcoatl and your troll account.
http://www.egyptsearch.com/forums/ultimatebb.cgi?ubb=get_topic;f=15;t=010625;p=4#000174
quote:That is totally false
Originally posted by xyyman:
[QB] Thanks Dr Winters and Q for bring this up. I wasn’t aware since 1999 it was confirmed that MtDNA Hg-M originated in Africa. And this was done the ‘right” way – through diversity analysis and NOT frequency.
![[Roll Eyes]](rolleyes.gif)
quote:Quetzalcoatl
In the Popol Vuh, the famous Mayan historian Ixtlilxochitl, the Olmecs came to Mexico in "ships of barks"(probably a reference to papyrus boats or dug-out canoes used by the Proto-Saharans) and landed in Potonchan, which they commenced to populate.Mexican traditions claim that these migrates from the east were led by Amoxaque or Bookmen. The term Amoxaque, is similar to the Manding 'a ma n'kye':"he (is) a teacher". These Blacks are frequently seen >in Mayan writings as gods or merchants.
quote:3) This passage in Ixtlilxochitl says nothing about the “amoxaque” WHICH IS NOT ANY KIND OF MAYA BUT NAHUATL.This term is found in Sahagun’s Florentine Codex. Winters is using Van Sertima or perhaps Wiener with his usual twist. Van Sertima argued that amoxaque really came from Egyptian [funny how pliable and flexible Nahuatl is- it resembles whatever language the current diffusionist needs (Shang Chinese, Egyptian, Mande, Phoenician, Latin, Welsh, etc.), whereas Winters says it is Mande. Both are full of baloney. To begin with 1) neither Van Sertima nor Winters knows enough to see that the word which they copied from the Spanish version not the Nahuatl version of the Florentine codex is misspelled; 2) Neither Van Sertima nor Winters knows that Nahuatl is an agglutinative language that elides letters so that the word they want to derive from either Egyptian or Mande is composed of AMOXTLI (“books”- HUA (possessive) QUE (plural form) to form AMOXHUAQUE pronounced /amoshwaque/ which has zero resemblance to Winters’ “so called” Mande which would need to be verified in any case given the track record we have seen already.
Those that possessed the new world in this third creation were the Olmecs and Xicalancas. According to the stories there are they came in ships or boats from the East to the land of Potonchan. Which they began to people. And on the shores of the Atoyac river which passes between the city of the Angels [the colonial city of Puebla] and Cholula [b(]this is near Mexico City not the Maya area[)/b] they met some of the giants who had escaped the catastrophe and extinction of the second creation of the earth. These giants being strong and trusting in their strength and size of body lorded it over the newcomers, in such a fashion that they oppressed them as if they were slaves . . .