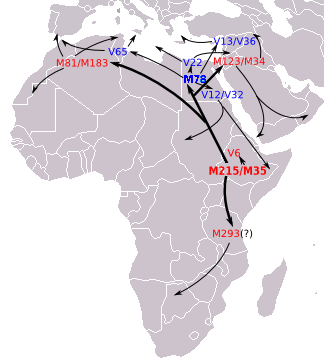
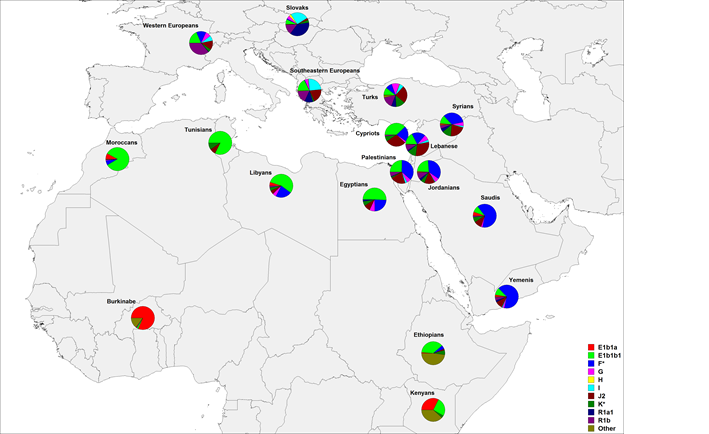
This large pre-Islamic inscription is depicted on a rock near a well in southern Arabia and consists of ten lines. It is popularly known as "the inscription of Abraha." The inscription is still in its original location; a replica is on display in the museum
نقش سبئي


(i)
Transliteration
b kh ya l / r h m n n / w m s ya h ha /
m l k n / a b r ha / z ya b m n / m l k / s b a / w z r ya d n / w h dh r m d t
Transcription
B'khail / ar-rahman / wmaseeha /
malikan / Abraha / Zaybm / malik /
sab'a / w zarydan / w hadarmaut
Translation
With the power (help) of god, and the Jesus (=Christian) King Abraha Zeebman (King's title), the King of Saba'a, Zuridan and Hadrmaut.
(ii)
w ya m n t / w r a a'in r b ha m r / ta w d m /
w t ha m t / s ta r w / z n / s ta r n / k gh z ya w
w yement / wa r'a rab hamw / Twadam / w thamat / satro / zn / satran / K'ghazow
and Yemen and the tribes (on)
the mountains and the coast wrote these lines on his battle
(iii)
m a'in d m / gh z w t n / r b a'in t n /
b w r kh n / z th b t n / k f s d w / k l / b n ya a'in m r m
Ma'ndam / Ghazwatn / rab'atan / b'warkhan / Zthbatan /Kafa saadu / kl/ bani amrm
against the tribe of Ma'ad ( in ) the battle of al-Rabiya in the
month of "Dhu al Thabithan" (April) and fight (against) all the (tribes) of Bani A'amir.
(iv)
w z k ya / m l k n / a b j b r / b a'in m /
k d t / w a'in l / w b sh r m / b n h sa n m / b a'in / m
Wazaki/ malikn/ abjabar / b ainam/ kadat/ wain/ w basharm / bin hasahanm/ bainm
and appointed the King (the leader) "Abi Jabar" with (tribe)
Kinda and (Qahtani tribe) Al (and the leader) "Bishar bin Hasan" with
(v)
s a'in d m / w m r d m / w h dh r w /
q d m ya / j ya sh n / a'in l ya b n ya a'in m r m /
k d t / w a'in l / b w d / z m r kh / w m r d m / w s d m / b w d.
San dam/ wa mardam / wa hadaru/ qadami / jayshan/
alia bani yamram/ kadat/ wail/ b wad /samrakh / wa mardam/ wa sadam/ b wad..
(Tribe) Sa'ad ( and the tribe) Murad and ( the tribe)
Hadarmaut (stand) in front of the army against Bani Amir of Kinda.
and (the tribe) Al in wadi "zu markh" and Murad and Sa'ad in wadi
(vi)
b m n ha j / t r b n / w z b h w / w a s r w /
w gh n m w / z a'in s m / w m kh dh / m l k n / b h l b n / w d n w.
B manhaj / tarban/ w zabahow / wa sarw /
w ghanamw / zaisam / wa makhdah/ malakin/ b halban/ wa danw
Manha on the way to Turban and killed and captured
and took the booty in large quantities and the
King and fought at Halban and reached
(vii)
k za l / m a'in d m / w r ha n w / w b a'in d n ha w /
w s a'in ha m w / a'in m r m / b n / m z r n..
Ka zalam/ maidam / wrahanw / wa badanahaw /
nwa sa'aham mw / amram / bin/ mazran.
Ma'ad and took booty and prisoners, and after that, conquered
(from the tribe of Ma'ad) Omro bin al-Munzir …
(viii)
w r ha n m w / b n ha w / w s t kh l fa ha w /
a'in l ya / m a'in d m / w q f l w / b n / h l
Wa rahanamw / bin haw / wa sata khalafw / ala/ ma'dam/ wa qafalw/ bin/ hal.
(and according to the agreement between Abrha and the tribe of Ma'ad)
(Abrhas) appointed the son (of Omro) as the ruler and returned (Abraha) from Hal.
(ix)
(b) n / (b) kh ya l / r h m n n / w r kh ha w /
z a'in l n / z l th n ya / w s th ya / w s
( bi)n / (b) akhayal / rahman / wa rakhaw / zalan / salthany / w sathya/ ws
Ban (halban) with the power of the god in the month of Zu A'allan in the year sixty-two
(x)
th / m a t m
Tha / matam
and six hundred
النص
ب خ ى ل / ر ح م ن ن / و م س ى ح هـ / م ل ك ن / أ ب ر هـ / ز ى ب م ن / م ل ك / س ب أ / و ذ ر ي د ن / و ح ض ر م و ت
القراءة
بقوة الرحمن ومسيحة الملك أبرهة زيبمان ملك سبأ وذو ريدان وحضرموت
ـ 2 ـ
و ي م ن ت / و ر أ ع ر ب هـ م و / ط و د م / و ت هـ م ت / س ط ر و / ذ ن / س ط ر ن / ك غ ز ى و
.ويمنات وقبائلهم (في) الجبال والسواحل ، سطر هذا النقش عندما غزا
ـ 3 ـ
م ع د م / غ ز و ت ن / ر ب ع ت ن / ب و ر خ ن / ذ ث ب ت ن / ك ف س د و / ك ل / ب ن ى ع م رم/
(قبيلة) معد (في) غزوة الربيع في شهر "ذو الثابة" (ابريل) عندما ثاروا كل (قبائل) بنى عامر
ـ 4 ـ
و ذ ك ى / م ل ك ن / أ ب ج ب ر / ب ع م / ك د ت / و ع ل / و ب ش ر م / ب ن ح ص ن م / ب ع م
وعين الملك (القائد) "أبي جبر" مع (قبيلة) على (والقائد) "بشر بن حصن" مع
ـ 5 ـ
س ع د م / و م ر د م / و ح ض ر و / ق د م ى / ج ي ش ن / ع ل ي / ب ن ي ع م ر م / ك د ت / و ع ل / ب و د / ذ م ر خ / و م ر د م / و س ع د م / ب و د
قبيلة) سعد (وقبيلة) مراد وحضروا أمام الجيش ـ ضد بنى عامر (وجهت) كندة وعلى في) وادي "ذو مرخ" ومراد وسعد في وادي
ـ 6 ـ
ب م ن هـ ج / ت ر ب ن / و ذ ب ح و / و أ س ر و / و غ ن م و / ذ ع س م / و م خ ض / م ل ك ن / ب ح ل ب ن / و د ن و
على طريق تربن وذبحوا وأسروا وغنموا بوفرة وحارب الملك في حلبن واقترب
ـ 7 ـ
ك ظ ل / م ع د م / و ر هـ ن و / و ب ع د ن هـ و / و س ع هـ م و / ع م ر م / ب ن / م ذ ر ن
كظل معد (وأخذ) اسرى، وبعد ذلك فوضوا (قبيلة معد) عمروا بن المنذر (في
ـ 8 ـ
و ر هـ ن هـ م و / ب ن هـ و / و س ت خ ل ف هـ و / ع ل ى / م ع د م / و ق ف ل و / ب ن / ح ل
الصلح) فضمنهم ابنه (عروا) (عن أبرهة) فعينه حاكماً على) معد ورجع (أبرهة) من حلـ
ـ 9 ـ
(ب) ن / ( ب ) خ ى ل / ر ح م ن ن / و ر خ هـ و / ذ ع ل ن / ذ ل ث ن ى / و س ث ى / و س
بن (حلبان) بقوة الرحمن في شهر ذو علان في السنة الثانية والستين وسـ
ـ 10 ـ
ث / م أ ت م
ستمائة
مسند جنوبي

Transliteration
ha z a'in
n b t a l
Transcription/Translation
Haza'a nabt al
(name of the deceased)
النص
ح ذ ع
ن ب ت أ ل
القراءة
حذع نبت أل
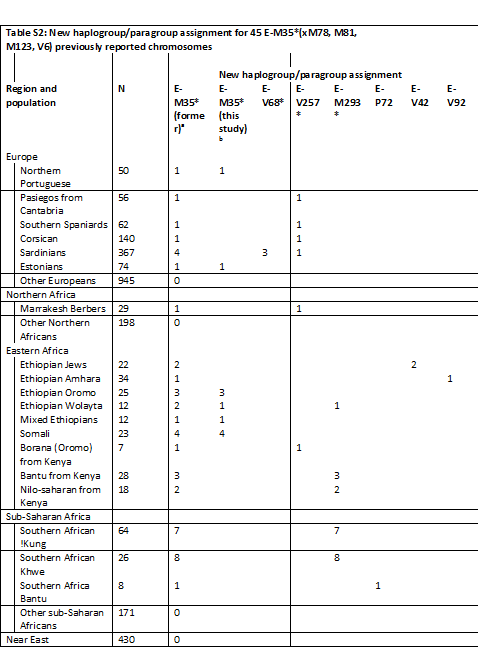
مسند جنوبي

Transliteration
n ya a'in th t / k ya l / w m q m / sh ya m ha m w
gh wa n ha m w / b n / a a'in r b n / w b z t
t a t b / r ya m m / s a'in d / w ha w f ya n
r ya m m / r dh w / w h sd ya / m r a ha m
Transcription
Nai Asath/ Khail/ w maqam/ shai mahamo/
Ghawnham/bin/ A'araban/ w bazat/
Ta'atab/remom/sad/w hawfain
Remom/ Rado/ wa hasiya/ mraham
Translation
With the power of Naiqthat and his high position
Ghawnaham from the Arabian tribe of
Dhat Ta'atab - Raimam Sa'ad
Fulfilled and pleased with the will of their Lord and his presence.
حجر عليه نقش مسند جنوبي مفقود جزء منه والجزء الواضح يتكون من اربعة أسطر كتبت بطريقة النقر وبخط غائر من اليمين إلى اليسار
النص
1- ن ي ع ث ت ، خ ي ل ، و م ق م ، ش ي م هـ م و ،
2- غ و ن هـ م و ، ب ن ، أ ع غ ب ن ، و ب ذ ت
3- ت أ ت ب ، ر ي م م ، س ع د ، و هـ و ف ي ن
4- ر ي م م ، ر ض و ، و ح ص ي ، م ر أ هـ م
القراءة
نيعست خيل ومقام شيمهمو
ونهمو بن (من) أعربن (بمعنى قبيلة) وبذت
(وتأتتب ريمم سعد وهوفين (بمعنى وأوفى
(ريمم بوصية المعبود رضو على مرآ هم (_على سمعهم





![[Big Grin]](biggrin.gif)
![[Wink]](wink.gif)

































![[Roll Eyes]](rolleyes.gif)




![[Confused]](confused.gif)
![[Embarrassed]](redface.gif)

![[Eek!]](eek.gif)




































![[Cool]](cool.gif)











































 .
.



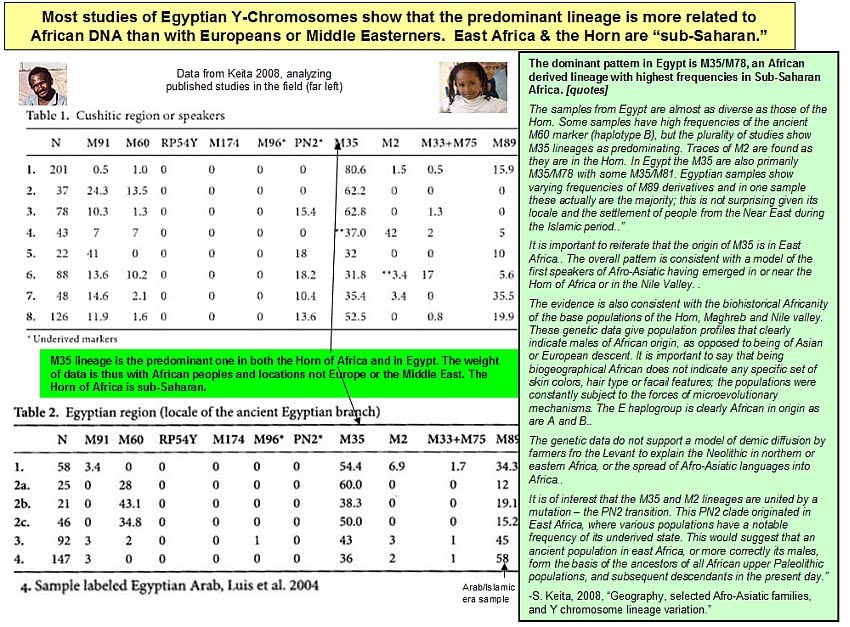
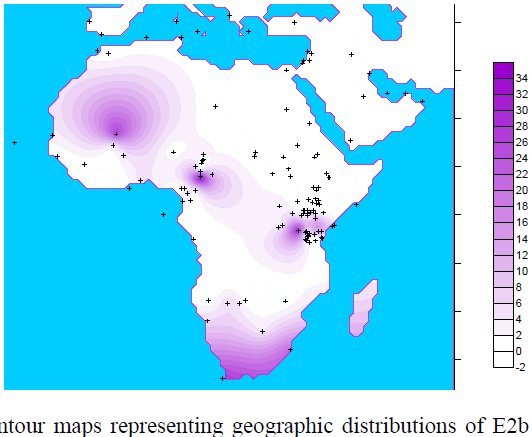
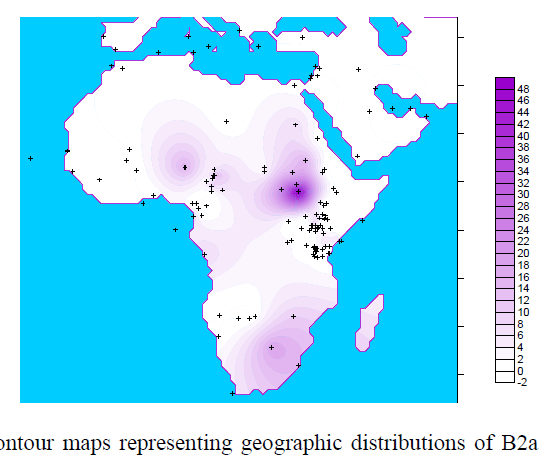
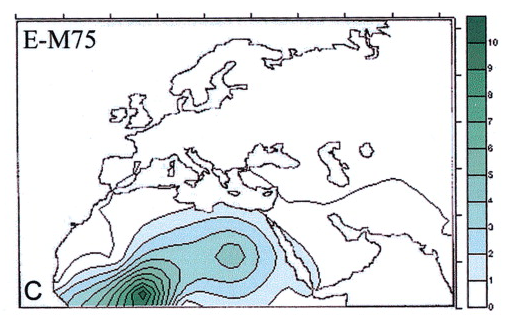














![[Razz]](tongue.gif) But, no! Dienekess did not have this one. Although my MO recently is for him to find and post and for me to critique. Much less work for me. He! He!
But, no! Dienekess did not have this one. Although my MO recently is for him to find and post and for me to critique. Much less work for me. He! He!